8F5T
 
 | | Rabbit muscle pyruvate kinase in complex with sodium and magnesium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F6M
 
 | | Complex of Rabbit muscle pyruvate kinase with ADP and the phosphonate analogue of PEP mimicking the Michaelis complex. | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, ADENOSINE-5'-DIPHOSPHATE, ALANINE, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
2F9G
 
 | | Crystal structure of Fus3 phosphorylated on Tyr182 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitogen-activated protein kinase FUS3 | | Authors: | Bhattacharyya, R.P, Remenyi, A, Good, M.C, Bashor, C.J, Falick, A.M, Lim, W.A. | | Deposit date: | 2005-12-05 | | Release date: | 2006-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway.
Science, 311, 2006
|
|
3I3A
 
 | | Structural Basis for the Sugar Nucleotide and Acyl Chain Selectivity of Leptospira interrogans LpxA | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, S-[2-({N-[(2R)-2-hydroxy-4-{[(S)-hydroxy(methoxy)phosphoryl]oxy}-3,3-dimethylbutanoyl]-beta-alanyl}amino)ethyl] (3R)-3-hydroxydodecanethioate | | Authors: | Williams, A.H. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA.
Biochemistry, 48, 2009
|
|
5OCV
 
 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | Deposit date: | 2017-07-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
3HT6
 
 | | 2-methylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | Lysozyme, PHOSPHATE ION, o-cresol | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTD
 
 | | (Z)-Thiophene-2-carboxaldoxime in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (NZ)-N-(thiophen-2-ylmethylidene)hydroxylamine, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
5NUC
 
 | | STAPHYLOCOCCAL NUCLEASE, 1-N-PENTANE THIOL DISULFIDE TO V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-11 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
1I2D
 
 | |
5GSX
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-2 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
4SGB
 
 | |
5NXH
 
 | |
8FLN
 
 | | Crystal structure of BTK C481S kinase domain in complex with pirtobrutinib | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
6TL3
 
 | | Crystal structure of an Estrogen Receptor alpha 8-mer phosphopeptide in complex with 14-3-3sigma stabilized by a Pyrrolidone1 derivative | | Descriptor: | 14-3-3 protein sigma, 5-[(2~{S},3~{R})-3-[(~{R})-azanyl(phenyl)methyl]-2-(4-nitrophenyl)-4,5-bis(oxidanylidene)pyrrolidin-1-yl]-2-oxidanyl-benzoic acid, Estrogen receptor | | Authors: | Andrei, S.A, Bosica, F, Ottmann, C, O'Mahony, G. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Design of Drug-Like Protein-Protein Interaction Stabilizers Guided By Chelation-Controlled Bioactive Conformation Stabilization.
Chemistry, 26, 2020
|
|
3KW3
 
 | |
4QX1
 
 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8FUL
 
 | |
8FUM
 
 | | AibH1H2 metalated with Fe in the presence of Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
8FK8
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
8F86
 
 | | SIRT6 bound to an H3K9Ac nucleosome | | Descriptor: | DNA (148-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Markert, J, Whedon, S, Wang, Z, Cole, P, Farnung, L. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.
J.Am.Chem.Soc., 145, 2023
|
|
8FUO
 
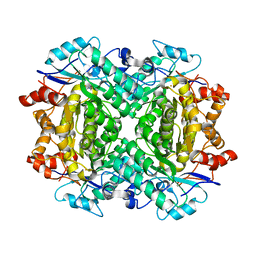 | | Fe-bound AibH1H2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, FE (III) ION | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
3HUK
 
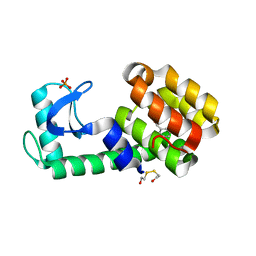 | | Benzylacetate in complex with T4 lysozyme L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
8FUN
 
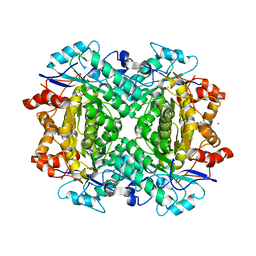 | | Enzymatically Active, Mn/Fe Metallated Form of AibH1H2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, FE (III) ION, ... | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
1ORF
 
 | | The Oligomeric Structure of Human Granzyme A Reveals the Molecular Determinants of Substrate Specificity | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Granzyme A, SULFATE ION | | Authors: | Bell, J.K, Goetz, D.H, Mahrus, S, Harris, J.L, Fletterick, R.J, Craik, C.S. | | Deposit date: | 2003-03-12 | | Release date: | 2003-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The oligomeric structure of human granzyme A is a determinant of its extended substrate specificity.
Nat.Struct.Biol., 10, 2003
|
|
3HO5
 
 | |
