7R1F
 
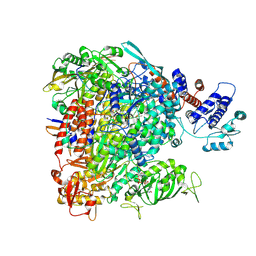 | |
7R0E
 
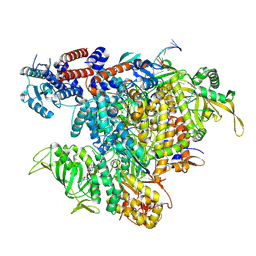 | |
6D90
 
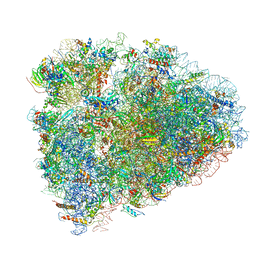 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-site tRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S12, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
7Q5B
 
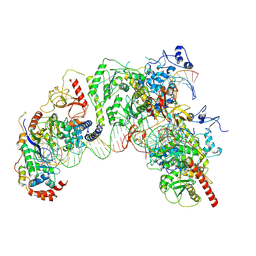 | | Cryo-EM structure of Ty3 retrotransposon targeting a TFIIIB-bound tRNA gene | | Descriptor: | DNA (19-MER), DNA (31-MER), DNA (34-MER), ... | | Authors: | Abascal-Palacios, G, Jochem, L, Pla-Prats, C, Beuron, F, Vannini, A. | | Deposit date: | 2021-11-03 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis of Ty3 retrotransposon integration at RNA Polymerase III-transcribed genes.
Nat Commun, 12, 2021
|
|
7UBM
 
 | |
7UBN
 
 | |
4Y2C
 
 | | M300V 3D polymerase mutant of EMCV | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
7KHE
 
 | | Escherichia coli RNA polymerase and rrnBP1 promoter pre-open complex with DksA/ppGpp | | Descriptor: | CHAPSO, DNA (46-MER), DNA (54-MER), ... | | Authors: | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
7KHI
 
 | | Escherichia coli RNA polymerase and rrnBP1 promoter complex with DksA/ppGpp | | Descriptor: | CHAPSO, DNA (28-MER), DNA (36-MER), ... | | Authors: | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2020-10-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
6PMI
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 1 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
6D9J
 
 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-sitetRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S12, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
6TYG
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 9 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TYF
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 6 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5X21
 
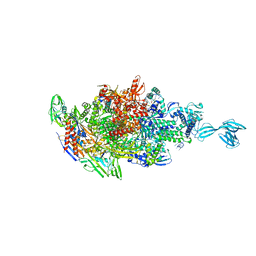 | | Crystal structure of Thermus thermophilus transcription initiation complex with GpA and pseudouridimycin (PUM) | | Descriptor: | (1S)-1,4-anhydro-5-[(N-carbamimidoylglycyl-N~2~-hydroxy-L-glutaminyl)amino]-5-deoxy-1-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zhang, Y, Ebright, R. | | Deposit date: | 2017-01-29 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase.
Cell, 169, 2017
|
|
5X22
 
 | |
7KHB
 
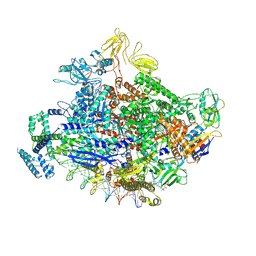 | | Escherichia coli RNA polymerase and rrnBP1 promoter open complex | | Descriptor: | CHAPSO, DNA (60-MER), DNA (64-MER), ... | | Authors: | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2020-10-20 | | Release date: | 2020-10-28 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
7KHC
 
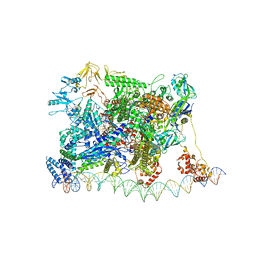 | | Escherichia coli RNA polymerase and rrnBP1 promoter closed complex | | Descriptor: | CHAPSO, DNA (18 MER), DNA (63-MER), ... | | Authors: | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2020-10-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
7DKH
 
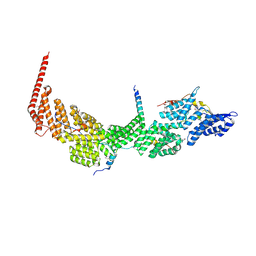 | | Crystal structure of the Ctr9/Paf1/Cdc73/Rtf1 quaternary complex | | Descriptor: | Cell division control protein 73, RNA polymerase II-associated protein 1, RNA polymerase-associated protein CTR9, ... | | Authors: | Chen, F.L, Liu, B.B, Guo, L, Li, D.F, Zhou, H, Long, J.F. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Core Module of the Yeast Paf1 Complex.
J.Mol.Biol., 434, 2021
|
|
6TYE
 
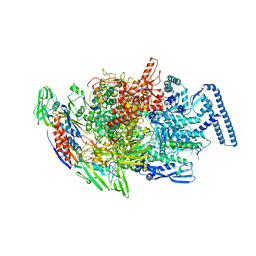 | | Crystal structure of MTB sigma L transcription initiation complex with 5 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RKP
 
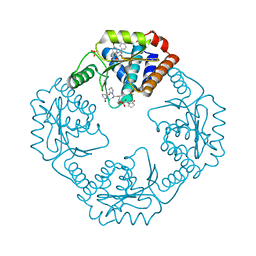 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with cyclic compound SJ001034733 | | Descriptor: | (5R)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2,6(28),11(27),12,14,22,24-octaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
3BSN
 
 | |
5ZVT
 
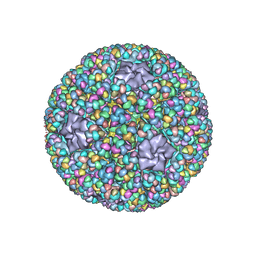 | | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly | | Descriptor: | C-terminus of outer capsid protein VP5, Core protein VP6, MYRISTIC ACID, ... | | Authors: | Liu, H, Fang, Q, Cheng, L. | | Deposit date: | 2018-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7K87
 
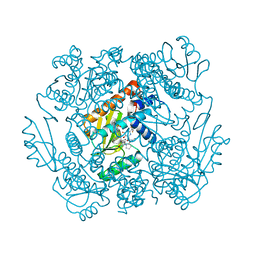 | | The crystal structure of the 2009 H1N1 PA endonuclease in complex with SJ000986436 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6Z9S
 
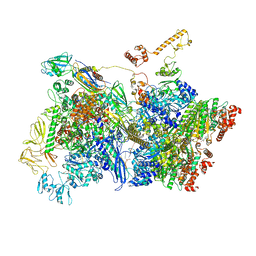 | | Transcription termination intermediate complex 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9R
 
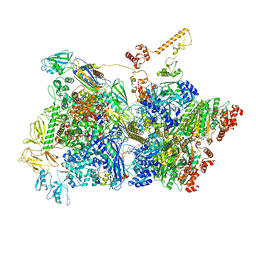 | | Transcription termination intermediate complex 3 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
