3GOZ
 
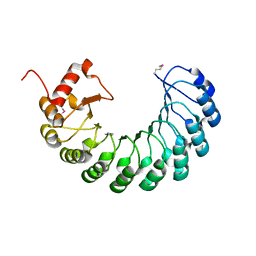 | | Crystal structure of the leucine-rich repeat-containing protein LegL7 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR148 | | Descriptor: | Leucine-rich repeat-containing protein | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Belote, R.L, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-20 | | Release date: | 2009-03-31 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of the leucine-rich repeat-containing protein LegL7 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR148.
To be Published
|
|
5OCG
 
 | | Discovery of small molecules binding to KRAS via high affinity antibody fragment competition method. | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, GTPase KRas, ... | | Authors: | Cruz-Migoni, A, Ehebauer, M.T, Phillips, S.E.V, Quevedo, C.E, Rabbitts, T.H. | | Deposit date: | 2017-06-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
7QPD
 
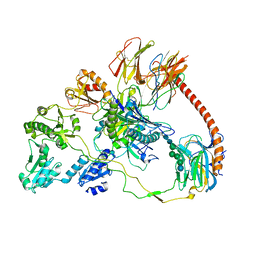 | | Structure of the human MHC I peptide-loading complex editing module | | Descriptor: | Beta-2-microglobulin, Calreticulin, HLA class I histocompatibility antigen, ... | | Authors: | Domnick, A, Susac, L, Trowitzsch, S, Thomas, C, Tampe, R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Molecular basis of MHC I quality control in the peptide loading complex.
Nat Commun, 13, 2022
|
|
6QJA
 
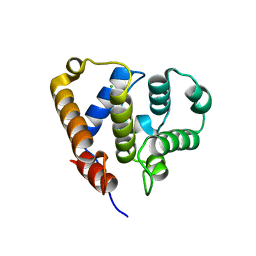 | | Organizational principles of the NuMA-Dynein interaction interface and implications for mitotic spindle functions | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nuclear mitotic apparatus protein 1 | | Authors: | Renna, C, Rizzelli, F, Carminati, M, Gaddoni, C, Pirovano, L, Cecatiello, V, Pasqualato, S, Mapelli, M. | | Deposit date: | 2019-01-23 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Organizational Principles of the NuMA-Dynein Interaction Interface and Implications for Mitotic Spindle Functions.
Structure, 28, 2020
|
|
7CND
 
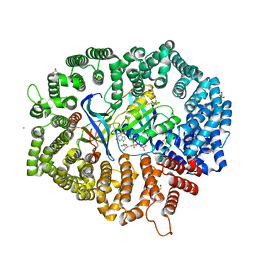 | | NCI-1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-07-31 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of the First Noncovalent Small-Molecule Inhibitor of CRM1.
J.Med.Chem., 64, 2021
|
|
5NXD
 
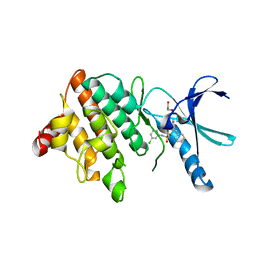 | | LIM Domain Kinase 2 (LIMK2) In Complex With TH-300 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-chlorophenyl)sulfamoyl]-~{N}-(phenylmethyl)-~{N}-propyl-benzamide, LIM domain kinase 2 | | Authors: | Mathea, S, Salah, E, Hanke, T, Newman, J.A, Oerum, S, Wang, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Knapp, S. | | Deposit date: | 2017-05-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | LIM Domain Kinase 2 (LIMK2)In Complex With TH-300
To Be Published
|
|
7COW
 
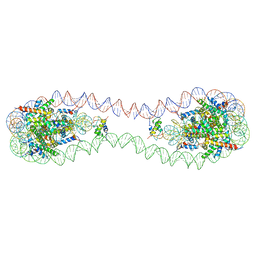 | | 353 bp di-nucleosome harboring cohesive DNA termini with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (353-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
5OB2
 
 | |
7QWJ
 
 | |
5M6C
 
 | | CRYSTAL STRUCTURE OF T71N MUTANT OF HUMAN HIPPOCALCIN | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Helassa, N, Antonyuk, S.V, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
3HP3
 
 | | Crystal structure of CXCL12 | | Descriptor: | CXCL12 protein, GOLD ION | | Authors: | Murphy, J.W, Lolis, E, Xiong, Y, Yuan, H, Crichlow, G. | | Deposit date: | 2009-06-03 | | Release date: | 2010-01-26 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heterologous quaternary structure of CXCL12 and its relationship to the CC chemokine family
Proteins, 78, 2009
|
|
5O8K
 
 | |
5OAV
 
 | |
7ROV
 
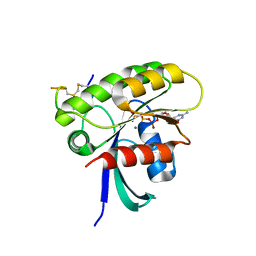 | | KRAS G12D Mutant in complex with GMPPCP and cyclic peptide MP-9903 | | Descriptor: | Cyclic peptide MP-9903, GLYCEROL, Isoform 2B of GTPase KRas, ... | | Authors: | Orth, P. | | Deposit date: | 2021-08-02 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of cell active macrocyclic peptides with on-target inhibition of KRAS signaling.
Chem Sci, 12, 2021
|
|
7RSE
 
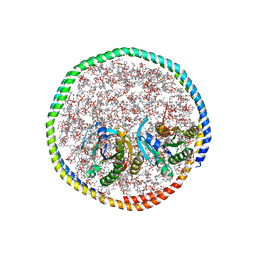 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSC
 
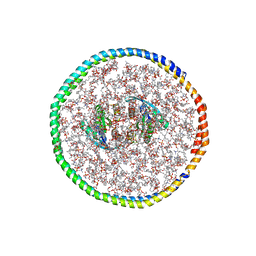 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7DBG
 
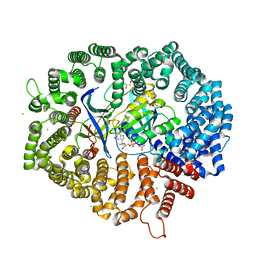 | | Yeast CRM1e (apo) in complex with Ran-RanBP1 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-10-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
7R04
 
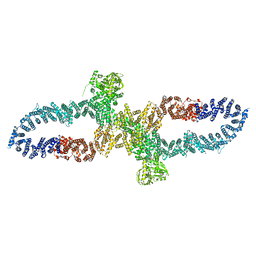 | | Neurofibromin in open conformation | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform I of Neurofibromin | | Authors: | Chaker-Margot, M, Scheffzek, K, Maier, T. | | Deposit date: | 2022-02-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of activation of the tumor suppressor protein neurofibromin.
Mol.Cell, 82, 2022
|
|
7R03
 
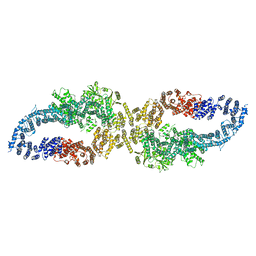 | |
7DN9
 
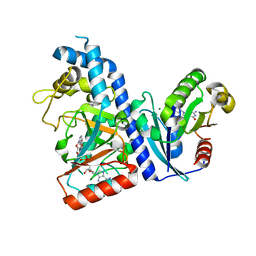 | |
7DN8
 
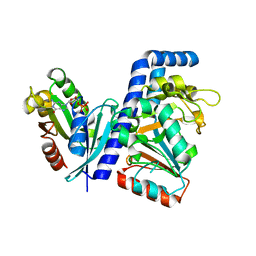 | | Crystal structure of Salmonella effector SopF in complex with ARF1 | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2020-12-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6084 Å) | | Cite: | ARF GTPases activate Salmonella effector SopF to ADP-ribosylate host V-ATPase and inhibit endomembrane damage-induced autophagy.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R24
 
 | |
7R5K
 
 | | Human nuclear pore complex (constricted) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Science, 376, 2022
|
|
7R5J
 
 | | Human nuclear pore complex (dilated) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (50 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores
Science, 376, 2022
|
|
7R4H
 
 | | phospho-STING binding to adaptor protein complex-1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Xu, P, Ablasser, A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Clathrin-associated AP-1 controls termination of STING signalling.
Nature, 610, 2022
|
|
