8GPC
 
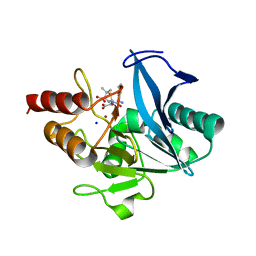 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, SODIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
2O36
 
 | |
2OHU
 
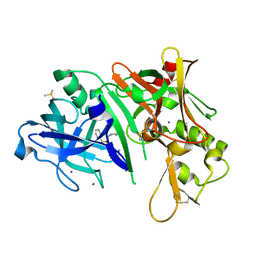 | | X-ray crystal structure of beta secretase complexed with compound 8b | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHT
 
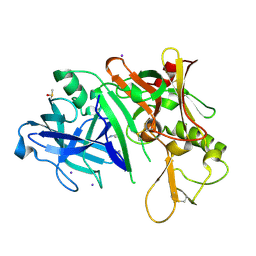 | | X-ray crystal structure of beta secretase complexed with compound 7 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
6Q37
 
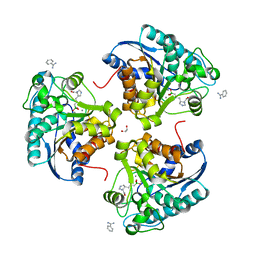 | | Complex of Arginase 2 with Example 23 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
2OKJ
 
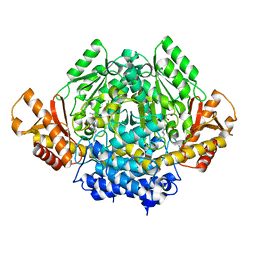 | | The X-ray crystal structure of the 67kDa isoform of Glutamic Acid Decarboxylase (GAD67) | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase 1 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8U0J
 
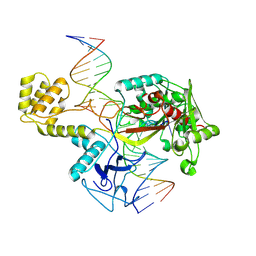 | | DdmE in complex with guide and target DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*TP*AP*AP*AP*GP*TP*CP*TP*AP*AP*CP*CP*TP*AP*TP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*CP*TP*AP*TP*AP*GP*GP*A)-3'), DNA (5'-D(P*GP*TP*TP*AP*GP*AP*CP*TP*TP*TP*AP*AP*GP*T)-3'), ... | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-08-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
2OQD
 
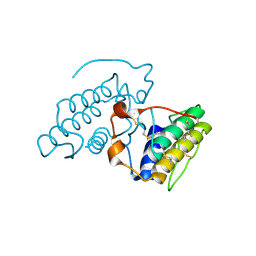 | | Crystal Structure of BthTX-II | | Descriptor: | Phospholipase A2 | | Authors: | Correa, L.C, Marchi-Salvador, D.P, Cintra, A.C.O, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of a myotoxic Asp49-phospholipase A(2) with low catalytic activity: Insights into Ca(2+)-independent catalytic mechanism.
Biochim.Biophys.Acta, 1784, 2008
|
|
2O3E
 
 | |
8U0W
 
 | | DdmD monomer in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-08-29 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
8U0U
 
 | | DdmD dimer in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-08-29 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
8G4Y
 
 | | Structure of ZNRF3 ECD bound to peptide MK1-3.6.10 | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, MK1-3.6.10 | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Potent and selective binders of the E3 ubiquitin ligase ZNRF3 stimulate Wnt signaling and intestinal organoid growth.
Cell Chem Biol, 31, 2024
|
|
8U3K
 
 | | DdmDE handover complex | | Descriptor: | DNA (29-MER), DNA (5'-D(P*GP*GP*AP*AP*AP*TP*GP*TP*TP*GP*AP*AP*TP*AP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
2OHS
 
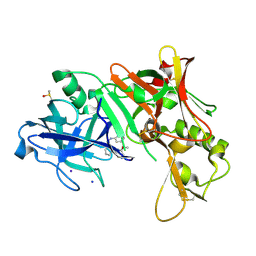 | | X-ray crystal structure of beta secretase complexed with compound 6b | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
3SNP
 
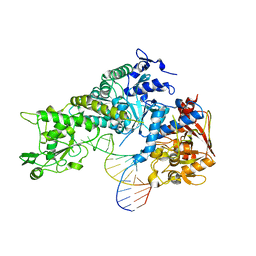 | |
3HNA
 
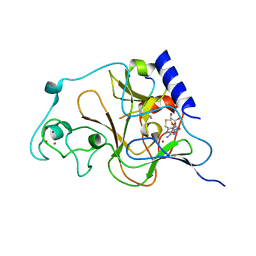 | | Crystal structure of catalytic domain of human euchromatic histone methyltransferase 1 in complex with SAH and mono-Methylated H3K9 Peptide | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, Mono-Methylated H3K9 Peptide, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Wleigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
3OIA
 
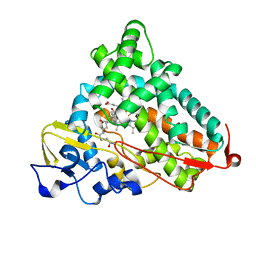 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8GluEtg-Bio | | Descriptor: | Camphor 5-monooxygenase, N-{15-oxo-19-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-4,7,10-trioxa-14-azanonadec-1-yl}-N'-(8-{[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbonyl]amino}octyl)pentanediamide, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8GluEtg-Bio
To be Published
|
|
2OHQ
 
 | | X-ray crystal structure of beta secretase complexed with compound 4 | | Descriptor: | 6-[2-(3'-METHOXYBIPHENYL-3-YL)ETHYL]PYRIDIN-2-AMINE, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
3P6X
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC3-C8-Dans | | Descriptor: | Camphor 5-monooxygenase, N-[8-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)octyl]-3-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]propanamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC3-C8-Dans
To be Published
|
|
3P6U
 
 | |
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
3P6V
 
 | |
8W2F
 
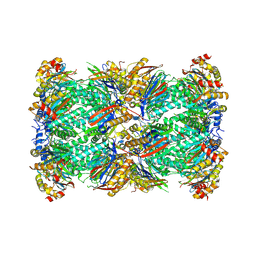 | | Plasmodium falciparum 20S proteasome bound to an inhibitor | | Descriptor: | (3S)-1-[(2-fluoroethoxy)acetyl]-N-{[(4P)-4-(6-methylpyridin-3-yl)-1,3-thiazol-2-yl]methyl}piperidine-3-carboxamide, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Han, Y, Deng, X, Ray, S, Chen, Z, Phillips, M. | | Deposit date: | 2024-02-20 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Identification of potent and reversible piperidine carboxamides that are species-selective orally active proteasome inhibitors to treat malaria.
Cell Chem Biol, 31, 2024
|
|
3P6S
 
 | |
2KMC
 
 | | Solution Structure of the N-terminal domain of kindlin-1 | | Descriptor: | Fermitin family homolog 1 | | Authors: | Goult, B.T, Bate, N, Roberts, G.C, Barsukov, I.L, Critchley, D.R. | | Deposit date: | 2009-07-27 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the N-Terminus of Kindlin-1: A Domain Important for alphaIIbbeta3 Integrin Activation
J.Mol.Biol., 394, 2009
|
|
