6ZVH
 
 | | EDF1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
6PV7
 
 | | Human alpha3beta4 nicotinic acetylcholine receptor in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gharpure, A, Teng, J, Zhuang, Y, Noviello, C.M, Walsh, R.M, Cabuco, R, Howard, R.J, Zaveri, N.T, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Agonist Selectivity and Ion Permeation in the alpha 3 beta 4 Ganglionic Nicotinic Receptor.
Neuron, 104, 2019
|
|
7MK5
 
 | | Crystal structure of Escherichia coli ClpP covalently inhibited by clipibicyclene | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-[(1E)-3-{[(2E,4E,6E,8S)-8-hydroxy-4-methyldeca-2,4,6-trienoyl]amino}-3-oxoprop-1-en-1-yl]azete-1(2H)-carboxylic acid, ACETATE ION, ... | | Authors: | Culp, E.J, Sychantha, D, Hobson, C, Pawlowski, A.J, Prehna, G, Wright, G.D. | | Deposit date: | 2021-04-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | ClpP inhibitors are produced by a widespread family of bacterial gene clusters.
Nat Microbiol, 7, 2022
|
|
7ABG
 
 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7A17
 
 | | Crystal structure of the 5-phosphatase domain of Synaptojanin1 bound to its substrate diC8-PI(3,4,5)P3 in complex with a nanobody | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, GLYCEROL, Isoform 2 of Synaptojanin-1, ... | | Authors: | Paesmans, J, Galicia, C, Martin, E, Versees, W. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A structure of substrate-bound Synaptojanin1 provides new insights in its mechanism and the effect of disease mutations.
Elife, 9, 2020
|
|
4YA2
 
 | |
8CYY
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B mixed type I/II with extended core from DLB case V | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8AS0
 
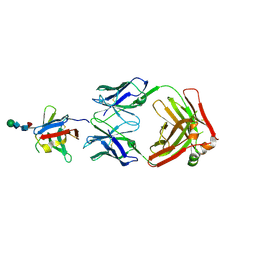 | | PD-1 extracellular domain in complex with Fab fragment from D12 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ongaro, T, Scietti, L, Pluss, L, Peissert, F, Villa, A, Puca, E, De Luca, R, Neri, D, Forneris, F. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Selection of a PD-1 blocking antibody from a novel fully human phage display library.
Protein Sci., 31, 2022
|
|
8QK9
 
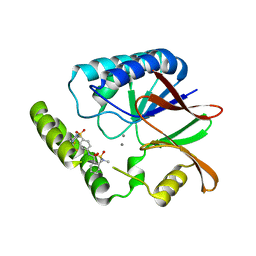 | | Structure of E. coli LpxH in complex with JEDI-1444 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-[4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]sulfonylphenyl]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4Y7W
 
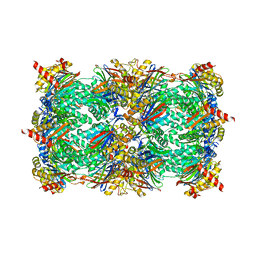 | | Yeast 20S proteasome in complex with Ac-LAE-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAE-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4YB6
 
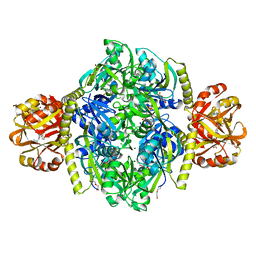 | | Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni in complex with the inhibitors AMP and histidine | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP phosphoribosyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mittelstaedt, G, Moggre, G.-J, Parker, E.J. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Campylobacter jejuni adenosine triphosphate phosphoribosyltransferase is an active hexamer that is allosterically controlled by the twisting of a regulatory tail.
Protein Sci., 25, 2016
|
|
6X8O
 
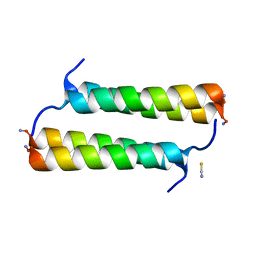 | | BimBH3 peptide tetramer | | Descriptor: | Bcl-2-like protein 11, THIOCYANATE ION | | Authors: | Robin, A.Y, Westphal, D, Uson, I, Czabotar, P.E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-09-09 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biophysical Characterization of Pro-apoptotic BimBH3 Peptides Reveals an Unexpected Capacity for Self-Association.
Structure, 29, 2021
|
|
8ITP
 
 | |
7QK0
 
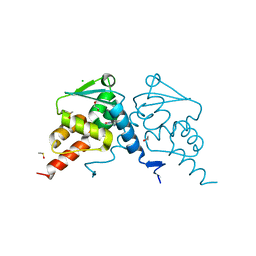 | | Crystal structure of human BCL6 BTB domain in complex with compound 12a | | Descriptor: | (2~{S})-10-[[5-chloranyl-2-[(3~{S},5~{R})-3-methyl-5-oxidanyl-piperidin-1-yl]pyrimidin-4-yl]amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Gunnell, E.A, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Improved Binding Affinity and Pharmacokinetics Enable Sustained Degradation of BCL6 In Vivo .
J.Med.Chem., 65, 2022
|
|
4Y8I
 
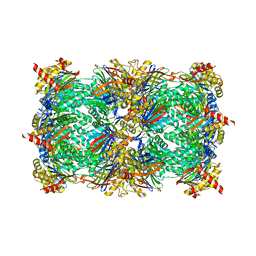 | | Yeast 20S proteasome in complex with Ac-PLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PLL-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6XSU
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens | | Descriptor: | ETHANOL, GH5-4 broad specificity endoglucanase | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
4YA9
 
 | |
6XRK
 
 | | GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-13 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6XSO
 
 | | GH5-4 broad specificity endoglucanase from an uncultured bacterium | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Cellulase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
8ITN
 
 | | Crystal structure of USP47apo catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 47, ZINC ION | | Authors: | Kim, E.E, Shin, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of USP47 reveals a hot spot for inhibitor design.
Commun Biol, 6, 2023
|
|
5EEA
 
 | | Structure of HOXB13-DNA(CAA) complex | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
8B7Y
 
 | | Cryo-EM structure of the E.coli 70S ribosome in complex with the antibiotic Myxovalargin B. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Koller, T.O, Graf, M, Wilson, D.N. | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Myxobacterial Antibiotic Myxovalargin: Biosynthesis, Structural Revision, Total Synthesis, and Molecular Characterization of Ribosomal Inhibition.
J.Am.Chem.Soc., 145, 2023
|
|
6YH7
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 3-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YH5
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 2-Chloro-4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-chloro-4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
7QYR
 
 | | Crystal structure of RimK from Pseudomonas aeruginosa PAO1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase, poly-glutamate | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
