6CGZ
 
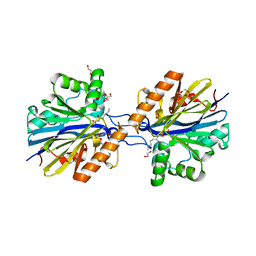 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to C6-AHL | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
5CFM
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' cGAMP, c[G(3', 5')pA(3', 5')p] | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CITRATE ANION, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
8JJR
 
 | | Cryo-EM structure of Symbiodinium photosystem I | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, L.S, Wang, N, Li, K, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of symbiotic dinoflagellate photosystem I-light-harvesting supercomplex in Symbiodinium.
Nat Commun, 15, 2024
|
|
4GZ6
 
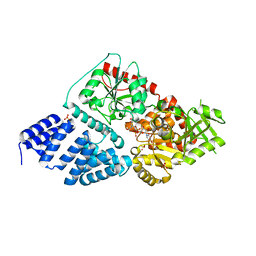 | | Crystal structure of human O-GlcNAc Transferase with UDP-5SGlcNAc | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
4H54
 
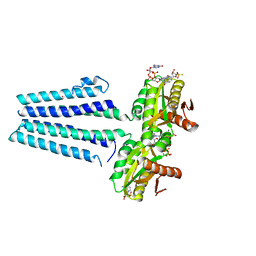 | | Crystal structure of the diguanylate cyclase DgcZ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Diguanylate cyclase YdeH, GUANOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, ... | | Authors: | Zaehringer, F, Schirmer, T. | | Deposit date: | 2012-09-18 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure and signaling mechanism of a zinc-sensory diguanylate cyclase.
Structure, 21, 2013
|
|
4H7U
 
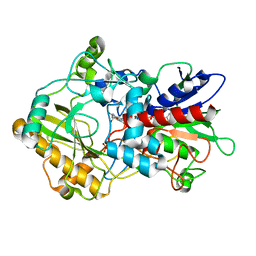 | | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Tan, T.C, Spadiut, O, Divne, C. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of pyranose dehydrogenase from Agaricus meleagris rationalizes substrate specificity and reveals a flavin intermediate.
Plos One, 8, 2013
|
|
4A33
 
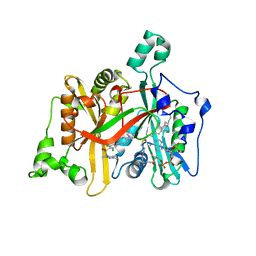 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 2,6-DICHLORO-4-(6-PIPERAZIN-1-YLPYRIDIN-3-YL)-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
4A30
 
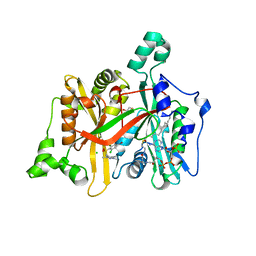 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-BROMO-2,6-DICHLORO-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
5WQU
 
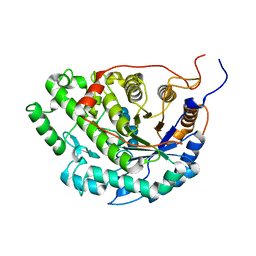 | | Crystal structure of Sweet Potato Beta-Amylase complexed with Maltotetraose | | Descriptor: | Beta-amylase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
6GCC
 
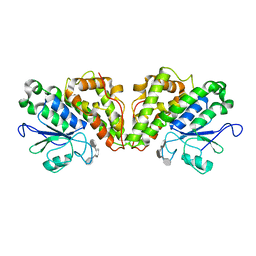 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor in complex with dextran-sulfate | | Descriptor: | 3,4-di-O-sulfo-alpha-D-glucopyranose-(1-6)-3,4-di-O-sulfo-alpha-D-altropyranose-(1-6)-1,2,3,4-tetra-O-sulfo-alpha-D-allopyranose, Glutathione transferase Xi 3 mutant C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-04-17 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
6GKV
 
 | | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | | Descriptor: | 6,7-dimethoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-2-ium, Coclaurine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dunstan, M.S, Levy, C.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Biocatalytic Scope of Coclaurine N-Methyltransferase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5CFN
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3',3' c-di-AMP, c[A(3',5')pA(3',5')p] | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
5TGY
 
 | | NMR structure of holo-PS1 | | Descriptor: | PS1, [5,10,15,20-tetrakis(trifluoromethyl)porphyrinato(2-)-kappa~4~N~21~,N~22~,N~23~,N~24~]zinc | | Authors: | Polizzi, N.F, Wu, Y. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
4IN6
 
 | | (M)L214A mutant of the Rhodobacter sphaeroides Reaction Center | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Saer, R.G, Hardjasa, A, Murphy, M.E.P, Beatty, J.T. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Role of Rhodobacter sphaeroides Photosynthetic Reaction Center Residue M214 in the Composition, Absorbance Properties, and Conformations of HA and BA Cofactors.
Biochemistry, 52, 2013
|
|
5BZS
 
 | |
4IN7
 
 | | (M)L214N mutant of the Rhodobacter sphaeroides Reaction Center | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Saer, R.G, Hardjasa, A, Murphy, M.E.P, Beatty, J.T. | | Deposit date: | 2013-01-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Role of Rhodobacter sphaeroides photosynthetic reaction center residue M214 in the composition, absorbance properties, and conformations of H(A) and B(A) cofactors.
Biochemistry, 52, 2013
|
|
3KXG
 
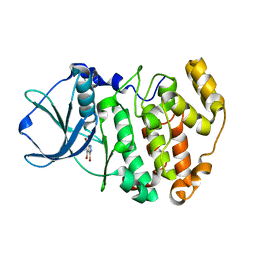 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor 3,4,5,6,7-pentabromo-1H-indazole (K64) | | Descriptor: | 3,4,5,6,7-pentabromo-1H-indazole, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
5C48
 
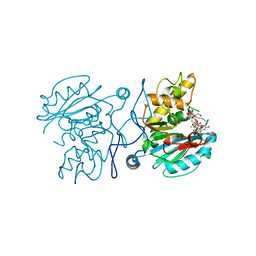 | | Crystal structure of GTB + UDP-C-Gal + DI | | Descriptor: | (((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)methyl)phosphonic (((2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Gagnon, S, Meloncelli, P, Zheng, R.B, Haji-Ghassemi, O, Johal, A.R, Borisova, S, Lowary, T.L, Evans, S.V. | | Deposit date: | 2015-06-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | High Resolution Structures of the Human ABO(H) Blood Group Enzymes in Complex with Donor Analogs Reveal That the Enzymes Utilize Multiple Donor Conformations to Bind Substrates in a Stepwise Manner.
J.Biol.Chem., 290, 2015
|
|
5U9D
 
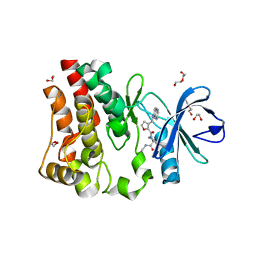 | | Discovery of a potent BTK inhibitor with a novel binding mode using parallel selections with a DNA-encoded chemical library | | Descriptor: | (R)-N-methyl-2-(3-((quinoxalin-6-ylamino)methyl)furan-2-carbonyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cuozzo, J.W, Centrella, P.A, Gikunju, D, Habeshian, S, Hupp, C.D, Keefe, A.D, Sigel, E, Soutter, H.H, Thomson, H.A, Zhang, Y, Clark, M.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library.
Chembiochem, 18, 2017
|
|
4NJI
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Mg2+ | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NMF
 
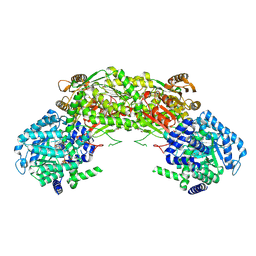 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA inactivated by N-propargylglycine and complexed with menadione bisulfite | | Descriptor: | (2R)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, (2S)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Singh, H, Tanner, J.J. | | Deposit date: | 2013-11-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5BNW
 
 | | The active site of O-GlcNAc transferase imposes constraints on substrate sequence | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, laminB1 residues 179-191 | | Authors: | Pathak, S, Alonso, J, Schimpl, M, Rafie, K, Blair, D.E, Borodkin, V.S, Albarbarawi, O, van Aalten, D.M.F. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4NJH
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxy-5,6,7,8-tetrahydropterin | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
3SWG
 
 | | AQUIFEX AEOLICUS MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys124 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
4A31
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 6-{[2-(4-METHYLPIPERAZIN-1-YL)ETHYL]AMINO}-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)PYRIDINE-3-SULFONAMIDE, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
