6PDZ
 
 | |
1ZNM
 
 | | A zinc finger with an artificial beta-turn, original sequence taken from the third zinc finger domain of the human transcriptional repressor protein YY1 (YING and YANG 1, a delta transcription factor), nmr, 34 structures | | Descriptor: | YY1, ZINC ION | | Authors: | Viles, J.H, Patel, S.U, Mitchell, J.B.O, Moody, C.M, Justice, D.E, Uppenbrink, J, Doyle, P.M, Harris, C.J, Sadler, P.J, Thornton, J.M. | | Deposit date: | 1997-11-20 | | Release date: | 1998-04-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Design, synthesis and structure of a zinc finger with an artificial beta-turn.
J.Mol.Biol., 279, 1998
|
|
8DKN
 
 | | PPARg bound to T0070907 and Co-R peptide | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1 peptide, ... | | Authors: | Larsen, N.A, Tsai, J. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DKV
 
 | | PPARg bound to JTP-426467 and Co-R peptide | | Descriptor: | 2-chloro-N-[4-(5-methyl-1,3-benzoxazol-2-yl)phenyl]-5-nitrobenzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
1JHC
 
 | | LEXA S119A C-TERMINAL TRYPTIC FRAGMENT | | Descriptor: | LEXA REPRESSOR | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JHE
 
 | | LEXA L89P Q92W E152A K156A MUTANT | | Descriptor: | LEXA REPRESSOR | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, J Strynadka, N.C. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1GDR
 
 | |
3V8K
 
 | |
3V8L
 
 | |
1MNM
 
 | |
7SXI
 
 | | Solution Structure of Sds3 Capped Tudor Domain | | Descriptor: | Sin3 histone deacetylase corepressor complex component SDS3 | | Authors: | Marcum, R.D, Radhakrishnan, I. | | Deposit date: | 2021-11-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A capped Tudor domain within a core subunit of the Sin3L/Rpd3L histone deacetylase complex binds to nucleic acid G-quadruplexes.
J.Biol.Chem., 298, 2022
|
|
8AQN
 
 | | Crystal structure of PPARG and NCOR2 with BAY-4931, an inverse agonist (compound 6c) | | Descriptor: | 2-chloranyl-~{N}-[2-(4-ethylphenyl)-1,3-benzoxazol-5-yl]-5-nitro-benzamide, CALCIUM ION, GLYCEROL, ... | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Braeuer, N, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Structure-Based Design of Potent Covalent PPAR gamma Inverse-Agonists BAY-4931 and BAY-0069 .
J.Med.Chem., 65, 2022
|
|
8AQM
 
 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 6a) | | Descriptor: | 2-chloranyl-~{N}-[2-(3-methylphenyl)-1,3-benzoxazol-5-yl]-5-nitro-benzamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Braeuer, N, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Structure-Based Design of Potent Covalent PPAR gamma Inverse-Agonists BAY-4931 and BAY-0069 .
J.Med.Chem., 65, 2022
|
|
3V8J
 
 | |
3H52
 
 | | Crystal structure of the antagonist form of human glucocorticoid receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Benz, J, D'Arcy, B, Stihle, M, Burger, D, Thoma, R, Ruf, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations
J.Mol.Biol., 395, 2010
|
|
2GPV
 
 | | Estrogen Related Receptor-gamma ligand binding domain complexed with 4-hydroxy-tamoxifen and a SMRT peptide | | Descriptor: | 4-HYDROXYTAMOXIFEN, Estrogen-related receptor gamma, Nuclear receptor corepressor 2 | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-18 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
6UXY
 
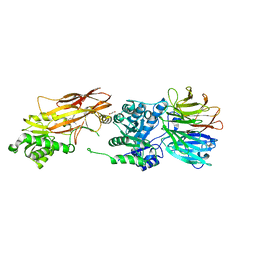 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 8 | | Descriptor: | (5R)-2-amino-5-(2-cyclohexylethyl)-3-methyl-5-phenyl-3,5-dihydro-4H-imidazol-4-one, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
6UXX
 
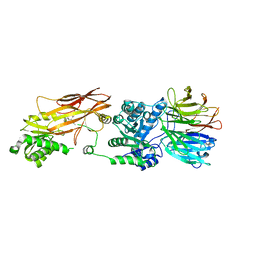 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 1a | | Descriptor: | (5R)-2-amino-5-(4-methoxyphenyl)-3-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-yl]-3,5-dihydro-4H-imidazol-4-one, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
6XYX
 
 | |
6XXS
 
 | |
1AHD
 
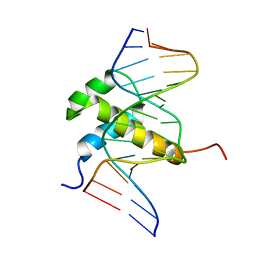 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF AN ANTENNAPEDIA HOMEODOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), Homeotic protein antennapedia | | Authors: | Billeter, M, Qian, Y.Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex.
J.Mol.Biol., 234, 1993
|
|
1FTZ
 
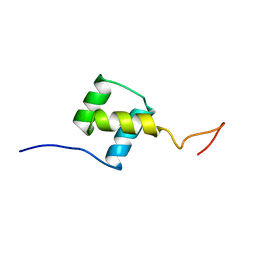 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE FUSHI TARAZU HOMEODOMAIN FROM DROSOPHILA AND COMPARISON WITH THE ANTENNAPEDIA HOMEODOMAIN | | Descriptor: | FUSHI TARAZU PROTEIN | | Authors: | Qian, Y.Q, Furukubo-Tokunaga, K, Resendez-Perez, D, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain.
J.Mol.Biol., 238, 1994
|
|
1MM3
 
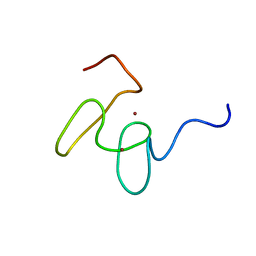 | | Solution structure of the 2nd PHD domain from Mi2b with C-terminal loop replaced by corresponding loop from WSTF | | Descriptor: | Mi2-beta(Chromodomain helicase-DNA-binding protein 4) and transcription factor WSTF, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
4BAY
 
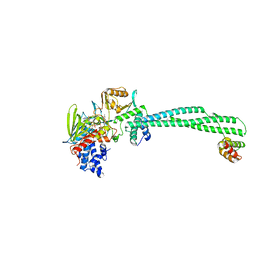 | | Phosphomimetic mutant of LSD1-8a splicing variant in complex with CoREST | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1 | | Authors: | Toffolo, E, Paganini, L, Rusconi, F, Tortorici, M, Pilotto, S, Verpelli, C, Tedeschi, G, Maffioli, E, Sala, C, Mattevi, A, Battaglioli, E. | | Deposit date: | 2012-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phosphorylation of Neuronal Lysine-Specific Demethylase 1Lsd1/Kdm1A Impairs Transcriptional Repression by Regulating Interaction with Corest and Histone Deacetylases Hdac1/2.
J.Neurochem., 128, 2014
|
|
5U50
 
 | |
