7PBC
 
 | | Crystal structure of engineered TCR (796) complexed to HLA-A*02:01 presenting MAGE-A10 9-mer peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
7PCK
 
 | |
3M62
 
 | |
1NBY
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K96A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
7PMU
 
 | | Crystal structure of native Iripin-8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, Serpin-8, ... | | Authors: | Polderdijk, S, Kotal, J, Chmelar, J, Huntington, J.A. | | Deposit date: | 2021-09-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Ixodes ricinus Salivary Serpin Iripin-8 Inhibits the Intrinsic Pathway of Coagulation and Complement.
Int J Mol Sci, 22, 2021
|
|
7PMY
 
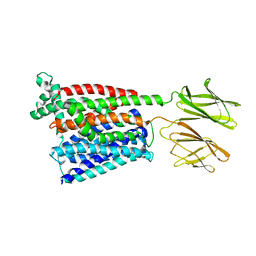 | | HsPepT2 bound to Ala-Phe in the inward facing partially occluded conformation | | Descriptor: | ALA-PHE, Solute carrier family 15 member 2 | | Authors: | Killer, M, Wald, J, Pieprzyk, J, Marlovits, T.C, Loew, C. | | Deposit date: | 2021-09-04 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes.
Sci Adv, 7, 2021
|
|
7PN1
 
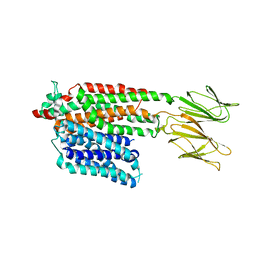 | | Apo HsPepT1 in the outward facing open conformation | | Descriptor: | Solute carrier family 15 member 1 | | Authors: | Killer, M, Wald, J, Pieprzyk, J, Marlovits, T.C, Loew, C. | | Deposit date: | 2021-09-04 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes.
Sci Adv, 7, 2021
|
|
7PMW
 
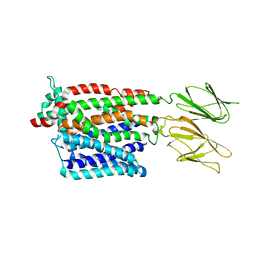 | | HsPepT1 bound to Ala-Phe in the outward facing occluded conformation | | Descriptor: | ALA-PHE, Solute carrier family 15 member 1 | | Authors: | Killer, M, Wald, J, Pieprzyk, J, Marlovits, T.C, Loew, C. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes.
Sci Adv, 7, 2021
|
|
7PMX
 
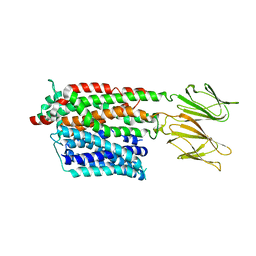 | | HsPepT1 bound to Ala-Phe in the outward facing open conformation | | Descriptor: | ALA-PHE, Solute carrier family 15 member 1 | | Authors: | Killer, M, Wald, J, Pieprzyk, J, Marlovits, T.C, Loew, C. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes.
Sci Adv, 7, 2021
|
|
3MOP
 
 | | The ternary Death Domain complex of MyD88, IRAK4, and IRAK2 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, Interleukin-1 receptor-associated kinase-like 2, Myeloid differentiation primary response protein MyD88 | | Authors: | Lin, S.-C, Lo, Y.-C, Wu, H. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling.
Nature, 465, 2010
|
|
7Q16
 
 | | Human 14-3-3 zeta fused to the BAD peptide including phosphoserine-74 | | Descriptor: | 14-3-3 protein zeta/delta,Bcl2-associated agonist of cell death | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Gushchin, I, Remeeva, A, Kovalev, K, Cooley, R.B. | | Deposit date: | 2021-10-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Crystal structure of human 14-3-3 zeta complexed with the noncanonical phosphopeptide from proapoptotic BAD.
Biochem.Biophys.Res.Commun., 583, 2021
|
|
3M9V
 
 | |
7PK9
 
 | | C-reactive protein decamer at pH 7.5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PM2
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, young JASP-stabilized F-actin, class 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
8I5K
 
 | |
8I5J
 
 | |
5C14
 
 | | Crystal structure of PECAM-1 D1D2 domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Zhou, D, Paddock, C, Newman, P, Zhu, J. | | Deposit date: | 2015-06-12 | | Release date: | 2016-01-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for PECAM-1 homophilic binding.
Blood, 127, 2016
|
|
1NBZ
 
 | | Crystal Structure of HyHEL-63 complexed with HEL mutant K97A | | Descriptor: | Lysozyme C, antibody kappa light chain, immunoglobulin gamma 1 chain | | Authors: | Mariuzza, R.A, Li, Y, Urrutia, M, Smith-Gill, S.J. | | Deposit date: | 2002-12-04 | | Release date: | 2003-04-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen
Biochemistry, 42, 2003
|
|
4MLM
 
 | | Crystal Structure of PhnZ from uncultured bacterium HF130_AEPn_1 | | Descriptor: | FE (III) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3D2O
 
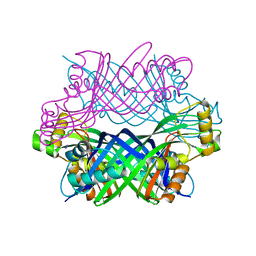 | | Crystal Structure of Manganese-metallated GTP Cyclohydrolase Type IB | | Descriptor: | AZIDE ION, CHLORIDE ION, LITHIUM ION, ... | | Authors: | Swairjo, M.A. | | Deposit date: | 2008-05-08 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB
J.Bacteriol., 191, 2009
|
|
4MLN
 
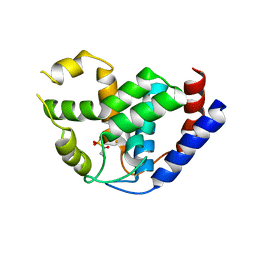 | | Crystal of PhnZ bound to (R)-2-amino-1-hydroxyethylphosphonic acid | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3UQ4
 
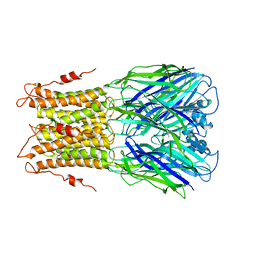 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant F247L (F16L) | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1, SODIUM ION | | Authors: | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | Deposit date: | 2011-11-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5AEC
 
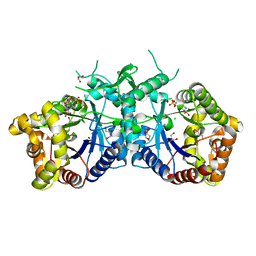 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8OF2
 
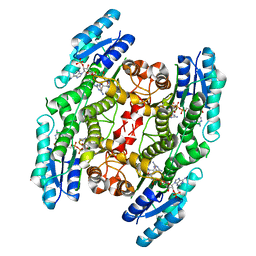 | | Trypanosoma brucei pteridine reductase 1 (TbPTR1) in complex with 2,4,6 triamminopyrimidine (TAP) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tassone, G, Landi, G, Mangani, S, Pozzi, C. | | Deposit date: | 2023-03-13 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The discovery of aryl-2-nitroethyl triamino pyrimidines as anti-Trypanosoma brucei agents.
Eur.J.Med.Chem., 264, 2023
|
|
5CUZ
 
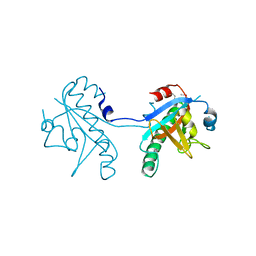 | | Crystal structure of SeMet-substituted N-terminal truncated human B12-chaperone CblD (108-296) | | Descriptor: | Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Yamada, K, Gherasim, C, Banerjee, R, Koutmos, M. | | Deposit date: | 2015-07-25 | | Release date: | 2015-09-23 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Human B12 Trafficking Protein CblD Reveals Molecular Mimicry and Identifies a New Subfamily of Nitro-FMN Reductases.
J.Biol.Chem., 290, 2015
|
|
