5MDV
 
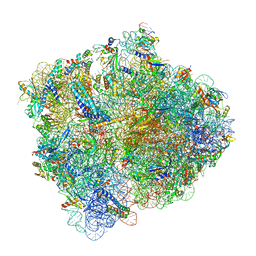 | | Structure of ArfA and RF2 bound to the 70S ribosome (accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5BZ7
 
 | |
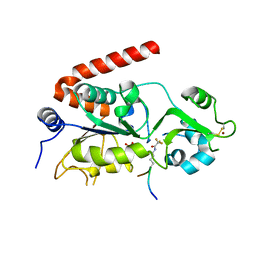
5FUO
 
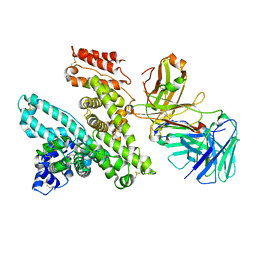 | |
5FYQ
 
 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
2D3D
 
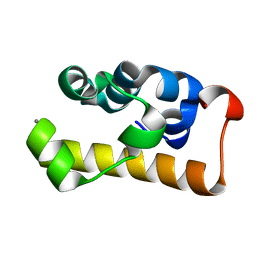 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
2D57
 
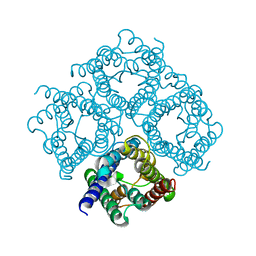 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
7RM8
 
 | |
2D8X
 
 | |
6B41
 
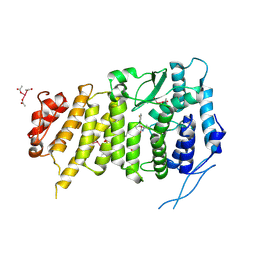 | | Menin bound to M-525 | | Descriptor: | Menin, methyl {(1S,2R)-2-[(S)-cyano[1-({1-[4-({1-[4-(dimethylamino)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl](3-fluorophenyl)methyl]cyclopentyl}carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design of the First-in-Class, Highly Potent Irreversible Inhibitor Targeting the Menin-MLL Protein-Protein Interaction.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1STC
 
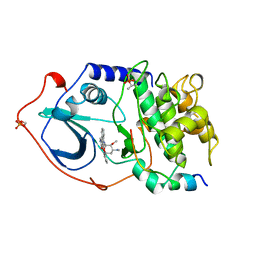 | | CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR, STAUROSPORINE | | Authors: | Prade, L, Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1997-10-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential.
Structure, 5, 1997
|
|
1SSK
 
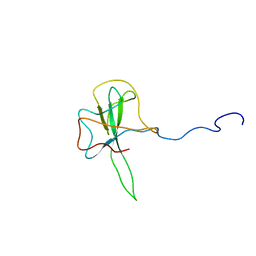 | | Structure of the N-terminal RNA-binding Domain of the SARS CoV Nucleocapsid Protein | | Descriptor: | Nucleocapsid protein | | Authors: | Huang, Q, Yu, L, Petros, A.M, Gunasekera, A, Liu, Z, Xu, N, Hajduk, P, Mack, J, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal RNA-Binding Domain of the SARS CoV Nucleocapsid Protein.
Biochemistry, 43, 2004
|
|
1SSP
 
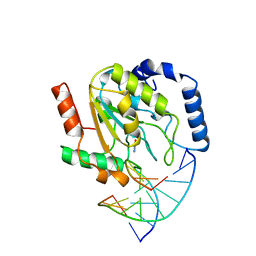 | | WILD-TYPE URACIL-DNA GLYCOSYLASE BOUND TO URACIL-CONTAINING DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*(D1P)P*AP*TP*CP*TP*T)-3', URACIL, ... | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
1SVG
 
 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 4 | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ALPHA FORM, N-{(3R,4R)-4-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZOYLAMINO]-AZEPAN-3-YL}ISONICOTINAMIDE, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
6B1O
 
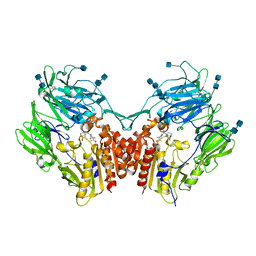 | | The structure of DPP4 in complex with Vildagliptin Analog | | Descriptor: | (2S)-2-amino-1-[(1S,3S,5S)-3-(aminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(1r,3R,5S,7S)-3,5-dihydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2017-09-18 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
6B76
 
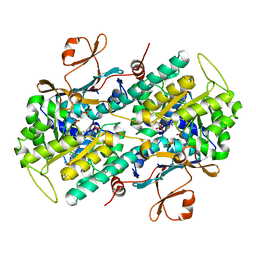 | | Crystal Structure of human NAMPT in complex with NVP-LVR596 | | Descriptor: | (1S,2S)-N-{4-[(1S)-1-(propanoylamino)ethyl]phenyl}-2-(pyridin-3-yl)cyclopropane-1-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
5BXW
 
 | |
1WNS
 
 | | Crystal structure of family B DNA polymerase from hyperthermophilic archaeon pyrococcus kodakaraensis KOD1 | | Descriptor: | DNA POLYMERASE | | Authors: | Hashimoto, H, Inoue, T, Kai, Y, Fujiwara, S, Takagi, M, Nishioka, M, Imanaka, T. | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-17 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DNA Polymerase from Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
J.Mol.Biol., 306, 2001
|
|
5MHR
 
 | | T3D reovirus sigma1 complexed with 9BG5 Fab fragments | | Descriptor: | 9BG5 Fab heavy chain, 9BG5 Fab light chain,LOC100046793 protein,MAb 110 light chain, Viral attachment protein sigma 1 | | Authors: | Stehle, T, Dietrich, M.H. | | Deposit date: | 2016-11-25 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into Reovirus sigma 1 Interactions with Two Neutralizing Antibodies.
J. Virol., 91, 2017
|
|
5GTU
 
 | |
1WIO
 
 | |
5BZ9
 
 | |
5MQF
 
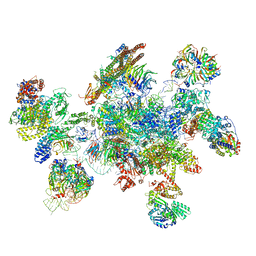 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | Authors: | Bertram, K, Hartmuth, K, Kastner, B. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
3DIG
 
 | |
5MRU
 
 | |
1T1U
 
 | | Structural Insights and Functional Implications of Choline Acetyltransferase | | Descriptor: | Choline O-acetyltransferase | | Authors: | Govindasamy, L, Pedersen, B, Lian, W, Kukar, T, Gu, Y, Jin, S, Agbandje-McKenna, M, Wu, D. | | Deposit date: | 2004-04-18 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights and functional implications of choline acetyltransferase
J.Struct.Biol., 148, 2004
|
|
