4N6T
 
 | | Adhiron: a stable and versatile peptide display scaffold - full length adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
4Q9Q
 
 | | Crystal structure of an RNA aptamer bound to bromo-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3-bromobenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4N6U
 
 | | Adhiron: a stable and versatile peptide display scaffold - truncated adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
9EQ3
 
 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The dynamics of hinge flexibility in receptor bound immunoglobulin E revealed by electron microscopy
Biorxiv, 2023
|
|
9BJM
 
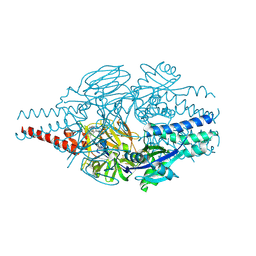 | | Crystal Structure of Inhibitor 5c in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1'-{[5-chloro-1-(4,4,4-trifluorobutyl)-1H-1,3-benzimidazol-2-yl]methyl}-1-(methanesulfonyl)spiro[azetidine-3,3'-indol]-2'(1'H)-one, Prefusion RSV F (DS-CAV1),Envelope glycoprotein | | Authors: | Shaffer, P.L, Milligan, C, Abeywickrema, P. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Spiro-Azetidine Oxindoles as Long-Acting Injectables for Pre-Exposure Prophylaxis against Respiratory Syncytial Virus Infections.
J.Med.Chem., 67, 2024
|
|
9EQ4
 
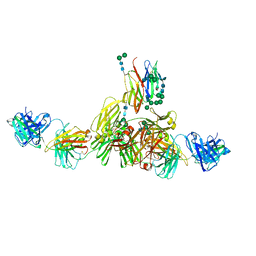 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The dynamics of hinge flexibility in receptor bound immunoglobulin E revealed by electron microscopy
Biorxiv, 2023
|
|
8ZGU
 
 | |
8Y81
 
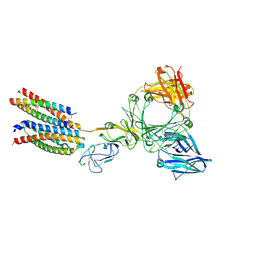 | | Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 633, 2024
|
|
8YM1
 
 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling.
J.Virol., 98, 2024
|
|
9CYL
 
 | | Structure of LAG3 loop1 deletion bound to the MHC class II molecule I-A(b) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Class-II-associated invariant chain peptide, H-2 class II histocompatibility antigen, ... | | Authors: | Ming, Q, Tran, T.H, Antfolk, D, Luca, V.C. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (4.66 Å) | | Cite: | Structural basis for mouse LAG3 interactions with the MHC class II molecule I-A b.
Nat Commun, 15, 2024
|
|
9EVO
 
 | |
9F9Y
 
 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
9EVN
 
 | |
9CYM
 
 | | Structure of LAG3 bound to the MHC class II molecule I-A(b) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Class-II-associated invariant chain peptide, H-2 class II histocompatibility antigen, ... | | Authors: | Ming, Q, Antfolk, D, Tran, T.H, Luca, V.C. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Structural basis for mouse LAG3 interactions with the MHC class II molecule I-A b.
Nat Commun, 15, 2024
|
|
8ZL9
 
 | | ASFV p72 in complex with Fab G6 | | Descriptor: | B646L, G6 Heavy chain, G6 Light chain | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-05-17 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
9FYD
 
 | | tubulin - cryptophycin-uD[Dab] complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dessin, C, Schachtsiek, T, Voss, J, Abel, A.-C, Neumann, B, Stammler, H.-G, Prota, A.E, Sewald, N. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Highly Cytotoxic Cryptophycin Derivatives with Modification in Unit D for Conjugation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8Y3Q
 
 | | ASFV p72 in complex with Fab F11 | | Descriptor: | B646L, Heavy chain of F11, Light chain of F11 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3O
 
 | | ASFV p72 in complex with Fab B1 | | Descriptor: | B646L, Heavy chain of B1, Light chain of B1 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3P
 
 | | ASFV p72 in complex with Fab C9 | | Descriptor: | B646L, Heavy chain of C9, Light chain of C9 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3R
 
 | | ASFV p72 in complex with Fab H3 | | Descriptor: | B646L, Heavy chain of H3, Light chain of H3 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
6OKD
 
 | | Crystal Structure of human transferrin receptor in complex with a cystine-dense peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Finton, K.A.K, Rupert, P.B, Strong, R.K. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A TfR-Binding Cystine-Dense Peptide Promotes Blood-Brain Barrier Penetration of Bioactive Molecules.
J.Mol.Biol., 432, 2020
|
|
6OKE
 
 | |
6P4J
 
 | | Mouse norovirus complexed with GCDCA | | Descriptor: | Capsid protein, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Smith, T.J, Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
6P4K
 
 | | mouse norovirus complexed with TCA | | Descriptor: | Capsid protein, TAUROCHOLIC ACID | | Authors: | Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
1BGS
 
 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | Descriptor: | BARNASE, BARSTAR | | Authors: | Guillet, V, Lapthorn, A, Mauguen, Y. | | Deposit date: | 1993-11-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
