5EJS
 
 | | Structure of Dictyostelium Discoideum Myosin VII MyTH4-FERM MF2 domain, mutant 1 | | Descriptor: | Myosin-I heavy chain | | Authors: | Planelles-Herrero, V.J, Sirkia, H, Sourigues, Y, Titus, M.A, Houdusse, A. | | Deposit date: | 2015-11-02 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin MyTH4-FERM structures highlight important principles of convergent evolution.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EOX
 
 | | Pseudomonas aeruginosa PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
7SD0
 
 | | Cryo-EM structure of the SHOC2:PP1C:MRAS complex | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liau, N.P.D, Johnson, M.C, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for SHOC2 modulation of RAS signalling.
Nature, 609, 2022
|
|
7SCW
 
 | | KRAS full length wild-type in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
7SCX
 
 | | KRAS full-length G12V in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
3F7Q
 
 | |
5F3X
 
 | | Crystal structure of Harmonin NPDZ1 in complex with ANKS4B SAM-PBM | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 4B, CHLORIDE ION, Harmonin | | Authors: | Li, J, He, Y, Lu, Q, Zhang, M. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex
Dev.Cell, 36, 2016
|
|
5F3Y
 
 | | Crystal Structure of Myo7b N-MyTH4-FERM-SH3 in complex with Anks4b CEN | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 4B, Unconventional myosin-VIIb | | Authors: | Li, J, He, Y, Lu, Q, Zhang, M. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex
Dev.Cell, 36, 2016
|
|
3D32
 
 | | Complex of GABA(A) receptor-associated protein (GABARAP) with a synthetic peptide | | Descriptor: | CHLORIDE ION, Gamma-aminobutyric acid receptor-associated protein, K1 peptide, ... | | Authors: | Weiergraeber, O.H, Stangler, T, Willbold, D. | | Deposit date: | 2008-05-09 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ligand Binding Mode of GABA(A) Receptor-Associated Protein.
J.Mol.Biol., 381, 2008
|
|
3CJ7
 
 | | Structure of Rattus norvegicus NTPDase2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ectonucleoside triphosphate diphosphohydrolase 2 | | Authors: | Zebisch, M, Strater, N. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6IXQ
 
 | | Structure of Myo2-GTD in complex with Smy1 | | Descriptor: | Kinesin-related protein SMY1, Myosin-2 | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
7SK6
 
 | | Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CID24 Fab heavy chain, CID24 Fab light chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK4
 
 | | Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK8
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK3
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK5
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12 and an intracellular Fab | | Descriptor: | Anti-Fab nanobody, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK7
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, and an extracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Anti-Fab nanobody, Atypical chemokine receptor 3, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7TD3
 
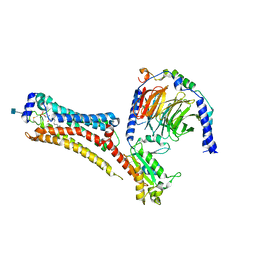 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD4
 
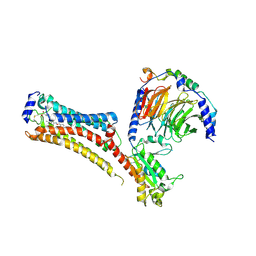 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to Siponimod | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7SWI
 
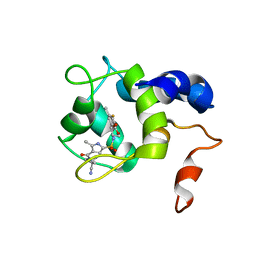 | | cTnC-TnI chimera complexed with A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SUP
 
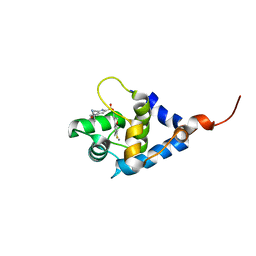 | | NMR structure of cTnC-TnI chimera bound to calcium and A1 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3S)-7-fluoro-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7T6B
 
 | | Structure of S1PR2-heterotrimeric G13 signaling complex | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, X, Chen, H. | | Deposit date: | 2021-12-13 | | Release date: | 2022-04-06 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of S1PR2-heterotrimeric G 13 signaling complex.
Sci Adv, 8, 2022
|
|
7SVC
 
 | | NMR structure of cTnC-TnI chimera bound to calcium and A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
6IXR
 
 | | Structure of Myo2-GTD in complex with Inp2 | | Descriptor: | Inheritance of peroxisomes protein 2, Myosin-2, SULFATE ION | | Authors: | Tang, K, Wei, Z. | | Deposit date: | 2018-12-11 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2.
J.Biol.Chem., 294, 2019
|
|
6HR2
 
 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
