5CXF
 
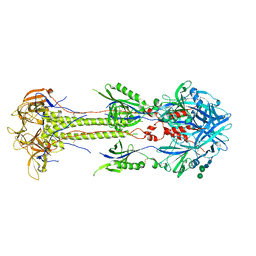 | |
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6U1N
 
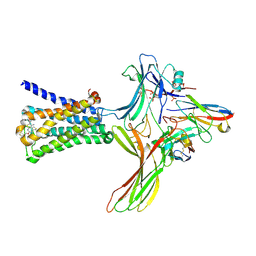 | | GPCR-Beta arrestin structure in lipid bilayer | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Staus, D.P, Hu, H, Robertson, M.J, Kleinhenz, A.L.W, Wingler, L.M, Capel, W.D, Latorraca, N.R, Lefkowitz, R.J, Skiniotis, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the M2 muscarinic receptor-beta-arrestin complex in a lipid nanodisc.
Nature, 579, 2020
|
|
6UMI
 
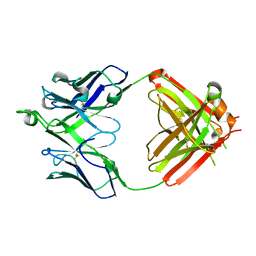 | | Crystal structure of erenumab Fab-b | | Descriptor: | erenumab Fab heavy chain, IgG1, erenumab Fab light chain | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMH
 
 | | Crystal structure of erenumab Fab-a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-PROPANDIOL, PHOSPHATE ION, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMJ
 
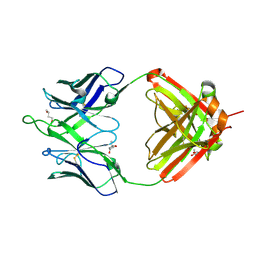 | | Crystal structure of erenumab Fab-c | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-BUTANEDIOL, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMG
 
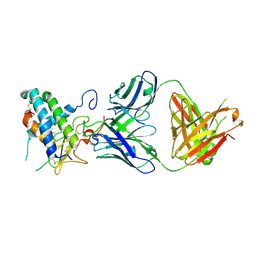 | | Crystal structure of erenumab Fab bound to the extracellular domain of CGRP receptor | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 1, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
8SFS
 
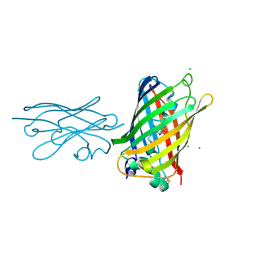 | | High Affinity nanobodies against GFP | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonnano, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SFZ
 
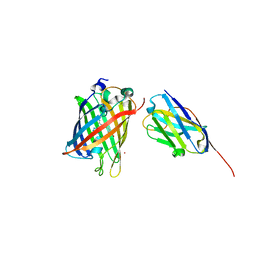 | | High Affinity nanobodies against GFP | | Descriptor: | Green fluorescent protein, LaG35, POTASSIUM ION, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
2CGR
 
 | |
8G0I
 
 | | High Affinity nanobodies against GFP | | Descriptor: | CHLORIDE ION, Green fluorescent protein, LaG24 Nanobody, ... | | Authors: | Ketaren, N.E, Rout, M.P, Almo, S. | | Deposit date: | 2023-01-31 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique Binding and Stabilization Mechanisms Employed By and Engineered Into Nanobodies
Biorxiv, 2023
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6ZDX
 
 | | RIFIN variable region bound to LILRB1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leukocyte immunoglobulin-like receptor subfamily B member 1, ... | | Authors: | Harrison, T.E, Higgins, M.K. | | Deposit date: | 2020-06-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RIFIN-mediated activation of LILRB1 in malaria.
Nature, 587, 2020
|
|
6OQ7
 
 | | Structure of the GTD domain of Clostridium difficile toxin B in complex with VHH E3 | | Descriptor: | E3, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8SFX
 
 | | High Affinity nanobodies against GFP | | Descriptor: | D-MALATE, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SFV
 
 | | High affinity nanobodies to GFP | | Descriptor: | GLYCEROL, Green fluorescent protein, LaG19, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High affinity nanobodies to GFP
To Be Published
|
|
8SG3
 
 | |
8SLC
 
 | | High Affinity nanobodies against GFP | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
2WQR
 
 | | The high resolution crystal structure of IgE Fc | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IG EPSILON CHAIN C REGION, ... | | Authors: | Dhaliwal, B, Sutton, B.J, Beavil, A.J. | | Deposit date: | 2009-08-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Changes in Ige Contribute to its Uniquely Slow Dissociation Rate from Receptor Fceri
Nat.Struct.Mol.Biol., 18, 2011
|
|
1CGS
 
 | |
8VQ9
 
 | |
8VQB
 
 | |
8VQA
 
 | |
5VN3
 
 | | Cryo-EM model of B41 SOSIP.664 in complex with soluble CD4 (D1-D2) and fragment antigen binding variable domain of 17b | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Pallesen, J, Ward, A.B. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike.
Nature, 547, 2017
|
|
6VFK
 
 | |
