4ZAE
 
 | | Development of a novel class of potent and selective FIXa inhibitors | | Descriptor: | 2,6-dichloro-N-[(2R)-2-(5,6-dimethyl-1H-benzimidazol-2-yl)-2-phenylethyl]-4-(4H-1,2,4-triazol-4-yl)benzamide, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, ... | | Authors: | Hruza, A, Reichert, P. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a novel class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6E04
 
 | | sperm whale myoglobin 1-nitrosopropane | | Descriptor: | 1-nitrosopropane, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
6I5M
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDP and formate ion | | Descriptor: | FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kravchenko, O, Nikonov, O, Gabdulkhakov, A, Stolboushkina, E, Arkhipova, V, Garber, M, Nikonov, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6IC1
 
 | | urate oxidase under 90 bar of krypton | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-AZAXANTHINE, ACETATE ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
4ZKY
 
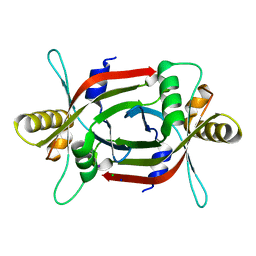 | | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, IODIDE ION, Pyridoxamine 5-phosphate oxidase, ... | | Authors: | Lee, B.M, Carr, P.D, Ahmed, F.H, Jackson, C.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
6I8Y
 
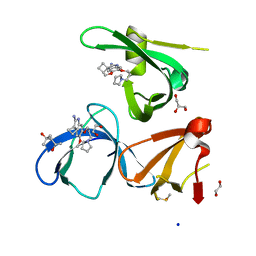 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I9X
 
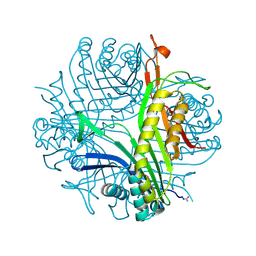 | | urate oxidase under 35 bar of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
4ZI8
 
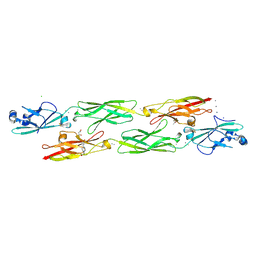 | | Structure of mouse clustered PcdhgC3 EC1-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Nicoludis, J.M, Lau, S.-Y, Scharfe, C.P.I, Marks, D.S, Weihofen, W.A, Gaudet, R. | | Deposit date: | 2015-04-27 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity.
Structure, 23, 2015
|
|
3QFD
 
 | |
6E8T
 
 | |
6EFC
 
 | | Hsa Siglec + Unique domains (unliganded) | | Descriptor: | CALCIUM ION, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
3HVI
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-ethyladenine-containing bisubstrate inhibitor | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
6EJC
 
 | |
5A10
 
 | | The crystal structure of Ta-TFP, a thiocyanate-forming protein involved in glucosinolate breakdown (space group C2) | | Descriptor: | ACETATE ION, SODIUM ION, THIOCYANATE FORMING PROTEIN | | Authors: | Krausze, J, Gumz, F, Wittstock, U. | | Deposit date: | 2015-04-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | The Crystal Structure of the Thiocyanate-Forming Protein from Thlaspi Arvense, a Kelch Protein Involved in Glucosinolate Breakdown.
Plant Mol.Biol., 89, 2015
|
|
6I3Z
 
 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin in complex with its antigen | | Descriptor: | Alpha-1-antitrypsin, Fab 2H2 heavy chain, Fab 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
3RH6
 
 | | DNA Polymerase Beta Mutant (Y271) with a dideoxy-terminated primer with an incoming ribonucleotide (rCTP) | | Descriptor: | (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DDG))-3'), (5'-D(P*GP*TP*CP*GP*G)-3'), 5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', ... | | Authors: | Cavanaugh, N.A, Beard, W.A, Batra, V.K, Perera, L, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2011-04-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Molecular insights into DNA polymerase deterrents for ribonucleotide insertion.
J.Biol.Chem., 286, 2011
|
|
3I58
 
 | |
1AVF
 
 | | ACTIVATION INTERMEDIATE 2 OF HUMAN GASTRICSIN FROM HUMAN STOMACH | | Descriptor: | GASTRICSIN, SODIUM ION | | Authors: | Khan, A.R, Cherney, M.M, Tarasova, N.I, James, M.N.G. | | Deposit date: | 1997-09-16 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural characterization of activation 'intermediate 2' on the pathway to human gastricsin.
Nat.Struct.Biol., 4, 1997
|
|
5A3W
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with Pyridine-2, 6-dicarboxylic Acid (PDCA) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
6EOM
 
 | | Structure of DPP III from Caldithrix abyssi | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, CHLORIDE ION, ... | | Authors: | Sabljic, I. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The first dipeptidyl peptidase III from a thermophile: Structural basis for thermal stability and reduced activity.
PLoS ONE, 13, 2018
|
|
3HUP
 
 | | High-resolution structure of the extracellular domain of human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69, SODIUM ION | | Authors: | Kolenko, P, Dohnalek, J, Skalova, T, Hasek, J, Duskova, J, Vanek, O, Bezouska, K. | | Deposit date: | 2009-06-15 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | The high-resolution structure of the extracellular domain of human CD69 using a novel polymer
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6I9Z
 
 | | urate oxidase under 65 bar of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
3I44
 
 | |
6ELE
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, fAB heavy chain, ... | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6EJE
 
 | |
