6NK6
 
 | | Electron Cryo-Microscopy Of Chikungunya VLP in complex with mouse Mxra8 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, E1 glycoprotein, ... | | Authors: | Basore, K, Kim, A.S, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM Structure of Chikungunya Virus in Complex with the Mxra8 Receptor.
Cell, 177, 2019
|
|
5MEN
 
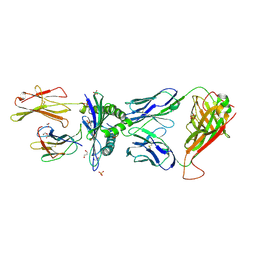 | | Human Leukocyte Antigen A02 presenting ILAKFLHWL, in complex with cognate T-Cell Receptor | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Lloyd, A, Crowther, M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Mechanism Underpinning Cross-reactivity of a CD8+ T-cell Clone That Recognizes a Peptide Derived from Human Telomerase Reverse Transcriptase.
J. Biol. Chem., 292, 2017
|
|
7TY0
 
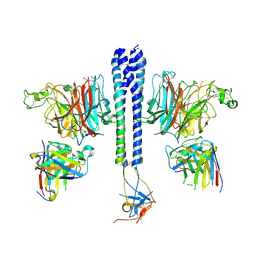 | |
7TJ8
 
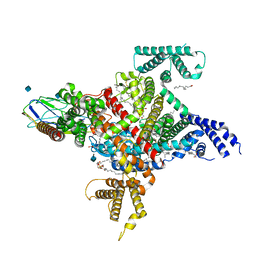 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in nanodiscs | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
7TJ9
 
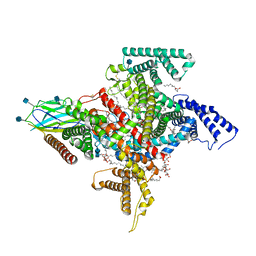 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in GDN | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
6NRW
 
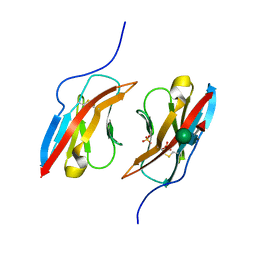 | | Crystal structure of Dpr1 IG1 bound to DIP-eta IG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
5MP3
 
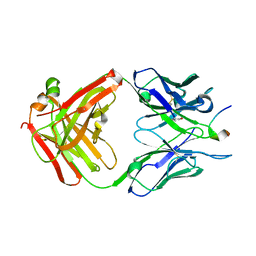 | |
6XDS
 
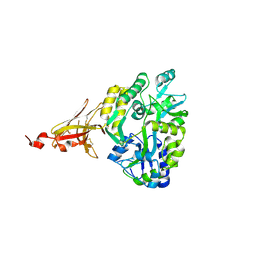 | |
6YJP
 
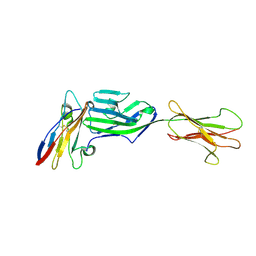 | | Crystal structure of a complex between glycosylated NKp30 and its deglycosylated tumour ligand B7-H6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Natural cytotoxicity triggering receptor 3, Natural cytotoxicity triggering receptor 3 ligand 1 | | Authors: | Skalova, T, Dohnalek, J, Skorepa, O, Kalouskova, B, Pazicky, S, Blaha, J, Vanek, O. | | Deposit date: | 2020-04-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Natural Killer Cell Activation Receptor NKp30 Oligomerization Depends on Its N -Glycosylation.
Cancers (Basel), 12, 2020
|
|
5MTL
 
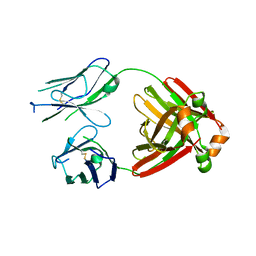 | | Crystal structure of an amyloidogenic light chain | | Descriptor: | light chain dimer,IGL@ protein,IGL@ protein | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
7UMG
 
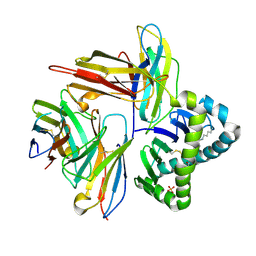 | | Crystal structure of human CD8aa-MR1-Ac-6-FP complex | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells.
J.Exp.Med., 219, 2022
|
|
5NHT
 
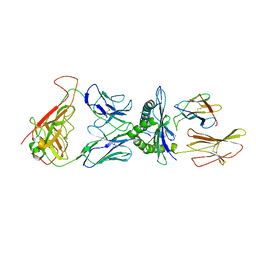 | | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
7VXZ
 
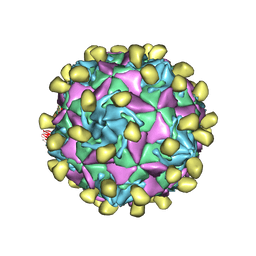 | |
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | Descriptor: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L, Ho, D.D. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-22 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
5NQK
 
 | | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-04-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
4BFI
 
 | | Structure of the complex of the extracellular portions of mouse CD200R and mouse CD200 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CELL SURFACE GLYCOPROTEIN CD200 RECEPTOR 1, ... | | Authors: | Hatherley, D, Lea, S.M, Johnson, S, Barclay, A.N. | | Deposit date: | 2013-03-19 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structures of Cd200/Cd200 Receptor Family and Implications for Topology, Regulation, and Evolution
Structure, 21, 2013
|
|
3ZL4
 
 | | Antibody structural organization: Role of kappa - lambda chain constant domain switch in catalytic functionality | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, A17 ANTIBODY FAB FRAGMENT HEAVY CHAIN, A17 ANTIBODY FAB FRAGMENT LAMBDA LIGHT CHAIN | | Authors: | Chatziefthimiou, S.D, Ponomarenko, N.A, Kurkova, I.N, Smirnov, A.V, Smirnov, I.V, Lamzin, V.S, Gabibov, A.G, Wilmanns, M. | | Deposit date: | 2013-01-28 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of Kappa>Lambda Light-Chain Constant-Domain Switch in the Structure and Functionality of A17 Reactibody
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CKD
 
 | |
4D9Q
 
 | |
4D9L
 
 | | Fab structure of anti-HIV-1 gp120 V2 mAb 697 | | Descriptor: | Heavy chain of Fab fragment of anti-HIV1 gp120 V2 mAb 697, Light chain of Fab fragment of anti-HIV1 gp120 V2 mAb 697 | | Authors: | Pan, R.M, Kong, X.P. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from HIV-1-infected individuals.
Virology, 427, 2012
|
|
8CWK
 
 | |
8E4C
 
 | | IgM BCR fab truncated form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain,Yellow fluorescent protein, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Dong, Y, Pi, X, Wu, H, Reth, M. | | Deposit date: | 2022-08-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural principles of B cell antigen receptor assembly.
Nature, 612, 2022
|
|
8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EDF
 
 | |
