2GPQ
 
 | |
2HB4
 
 | | Structure of HIV Protease NL4-3 in an Unliganded State | | Descriptor: | MAGNESIUM ION, Protease, R-1,2-PROPANEDIOL | | Authors: | Heaslet, H, Tam, K, Elder, J.H, Stout, C.D. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility in the flap domains of ligand-free HIV protease.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3WG2
 
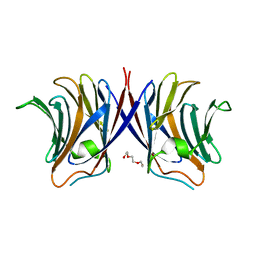 | | Crystal structure of Agrocybe cylindracea galectin mutant (N46A) | | Descriptor: | Galactoside-binding lectin, TRIETHYLENE GLYCOL | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
4DRF
 
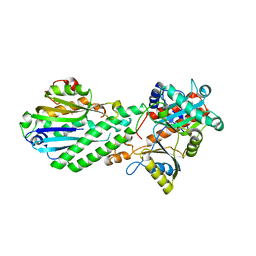 | | Crystal Structure of Bacterial Pnkp-C/Hen1-N Heterodimer | | Descriptor: | GLYCEROL, Metallophosphoesterase, Methyltransferase type 12 | | Authors: | Huang, R.H, Wang, P. | | Deposit date: | 2012-02-17 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of bacterial protein Hen1 activating the ligase activity of bacterial protein Pnkp for RNA repair.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2HM9
 
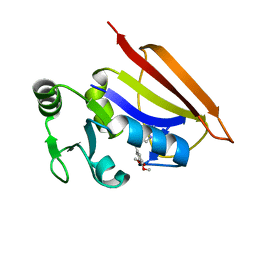 | | Solution structure of dihydrofolate reductase complexed with trimethoprim, 33 structures | | Descriptor: | 2,4-DIAMINO-5-(3,4,5-TRIMETHOXY-BENZYL)-PYRIMIDIN-1-IUM, Dihydrofolate reductase | | Authors: | Polshakov, V.I, Birdsall, B. | | Deposit date: | 2006-07-11 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structures of apo L.casei dihydrofolate reductase and its complexes with trimethoprim and NADPH. Contributions to positive cooperative binding from ligand-induced refolding, conformational changes and interligand hydrophobic interactions
Biochemistry, 2011
|
|
3WG4
 
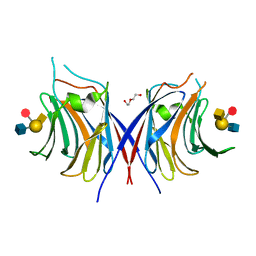 | | Crystal structure of Agrocybe cylindracea galectin mutant (N46A) with blood type A antigen tetraose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Galactoside-binding lectin, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
3WU2
 
 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Umena, Y, Kawakami, K, Shen, J.R, Kamiya, N. | | Deposit date: | 2014-04-21 | | Release date: | 2014-09-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A
Nature, 473, 2011
|
|
3WAQ
 
 | | Hemerythrin-like domain of DcrH I119E mutant (met) | | Descriptor: | Hemerythrin-like domain protein DcrH, MU-OXO-DIIRON | | Authors: | Okamoto, Y, Onoda, A, Sugimoto, H, Takano, Y, Hirota, S, Kurtz Jr, D.M, Shiro, Y, Hayashi, T. | | Deposit date: | 2013-05-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure, exogenous ligand binding, and redox properties of an engineered diiron active site in a bacterial hemerythrin
Inorg.Chem., 52, 2013
|
|
3WG3
 
 | | Crystal structure of Agrocybe cylindracea galectin with blood type A antigen tetraose | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Galactoside-binding lectin, ... | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
8V9O
 
 | | Imaging scaffold engineered to bind the therapeutic protein target BARD1 | | Descriptor: | CALCIUM ION, Tetrahedral Nanocage Cage Component Fused to Anti-BARD1 Darpin, Tetrahedral Nanocage Cage, ... | | Authors: | Agdanowski, M.P, Castells-Graells, R, Sawaya, M.R, Yeates, T.O, Arbing, M.A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | X-ray crystal structure of a designed rigidified imaging scaffold in the ligand-free conformation.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9EO0
 
 | | Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION, ~{N}-[3-[3-[[5-[(2-hydroxyethylamino)methyl]pyridin-2-yl]carbonylamino]-2-methyl-phenyl]-2-methyl-phenyl]-5-[[3-(methylsulfonylamino)propylamino]methyl]pyridine-2-carboxamide | | Authors: | Plewka, J, Hec, A, Sitar, T, Holak, T. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonsymmetrically Substituted 1,1'-Biphenyl-Based Small Molecule Inhibitors of the PD-1/PD-L1 Interaction.
Acs Med.Chem.Lett., 15, 2024
|
|
1F4F
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH SP-722 | | Descriptor: | 4-[[GLUTAMIC ACID]-CARBONYL]-BENZENE-SULFONYL-D-PROLINE, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4E6N
 
 | | Crystal structure of bacterial Pnkp-C/Hen1-N heterodimer | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Metallophosphoesterase, ... | | Authors: | Huang, R.H, Wang, P. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular basis of bacterial protein Hen1 activating the ligase activity of bacterial protein Pnkp for RNA repair.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8CM8
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with 4-(bromophenyl)phthalazinone D-galactal ligand | | Descriptor: | 4-(4-bromophenyl)-2-[[(2~{R},3~{R},4~{R})-2-(hydroxymethyl)-3-oxidanyl-3,4-dihydro-2~{H}-pyran-4-yl]oxymethyl]phthalazin-1-one, CHLORIDE ION, Galectin-8 | | Authors: | Van Klaveren, S, Hakansson, M, Diehl, C, Nilsson, N.J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Halogenated Galactal-Phenylphthalazinone Hybrids as Highly Selective Galectin-8N Ligands
To Be Published
|
|
3F1P
 
 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5OBH
 
 | | Crystal structure of glycine binding protein in complex with bicuculline | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dawson, A, Hunter, W.N, Jones, M. | | Deposit date: | 2017-06-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Structural Rationale for N-Methylbicuculline Acting as a Promiscuous Competitive Antagonist of Inhibitory Pentameric Ligand-Gated Ion Channels.
Chembiochem, 21, 2020
|
|
1F4B
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THYMIDYLATE SYNTHASE | | Descriptor: | GLYCEROL, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F4G
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH SP-876 | | Descriptor: | GLYCEROL, N-[4-[[GLUTAMIC ACID]-CARBONYL]-BENZENE-SULFONYL-D-PROLINYL]-3-AMINO-PROPANOIC ACID, SULFATE ION, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F4C
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COVALENTLY MODIFIED AT C146 WITH N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL | | Descriptor: | GLYCEROL, N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL, SULFATE ION, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F4E
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH TOSYL-D-PROLINE | | Descriptor: | GLYCEROL, SULFATE ION, THYMIDYLATE SYNTHASE, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4EOZ
 
 | |
3F1N
 
 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with internally bound ethylene glycol. | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4OYT
 
 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with D-serine and folinic acid | | Descriptor: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-oxidanyl-propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-02-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4APU
 
 | | PR X-Ray structures in agonist conformations reveal two different mechanisms for partial agonism in 11beta-substituted steroids | | Descriptor: | (8S,11R,13S,14S,16S,17S)-17-cyclopropylcarbonyl-16-ethenyl-13-methyl-11-(4-pyridin-3-ylphenyl)-2,6,7,8,11,12,14,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-one, 2-CHLORO-N-[[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]METHYL]-1,4-DIMETHYL-1H-PYRAZOLE-4-SULFONAMIDE, PROGESTERONE RECEPTOR, ... | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Bosch, R, Vu-Pham, D, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of progesterone receptor ligand binding domain in its agonist state reveal differing mechanisms for mixed profiles of 11 beta-substituted steroids.
J. Biol. Chem., 287, 2012
|
|
2IPO
 
 | | E. coli Aspartate Transcarbamoylase complexed with N-phosphonacetyl-L-asparagine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Cardia, J.P, Eldo, J, Xia, J, O'Day, E.M, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Use of L-asparagine and N-phosphonacetyl-L-asparagine to investigate the linkage of catalysis and homotropic cooperativity in E. coli aspartate transcarbomoylase.
Proteins, 71, 2008
|
|
