3JB9
 
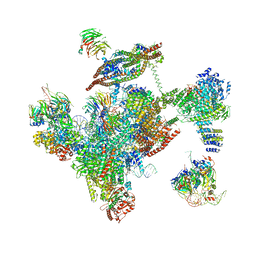 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
3JBA
 
 | | The U4 antibody epitope on human papillomavirus 16 identified by cryo-EM | | Descriptor: | H16.U4 antibody heavy chain, H16.U4 antibody light chain, Major capsid protein L1 | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Lee, H, Ashley, R.E, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | The U4 Antibody Epitope on Human Papillomavirus 16 Identified by Cryo-electron Microscopy.
J.Virol., 89, 2015
|
|
3JBB
 
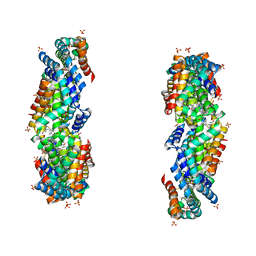 | | Characterization of red-shifted phycobiliprotein complexes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris | | Descriptor: | PHYCOCYANOBILIN, SULFATE ION, allophycocyanin beta chain, ... | | Authors: | Li, Y, Lin, Y, Garvey, C, Birch, D, Corkery, R.W, Loughlin, P.C, Scheer, H, Willows, R.D, Chen, M. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-11 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Characterization of red-shifted phycobilisomes isolated from the chlorophyll f-containing cyanobacterium Halomicronema hongdechloris.
Biochim.Biophys.Acta, 1857, 2015
|
|
3JBC
 
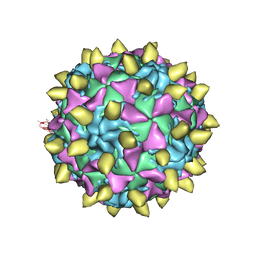 | | Complex of Poliovirus with VHH PVSP29F | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBD
 
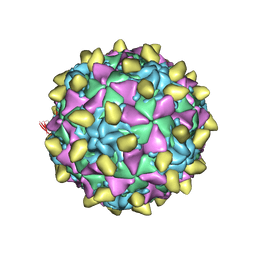 | | Complex of poliovirus with VHH PVSP6A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBE
 
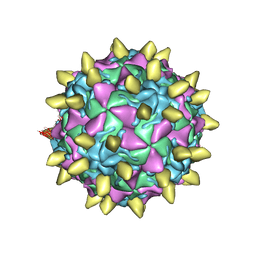 | | Complex of poliovirus with VHH PVSS8A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBF
 
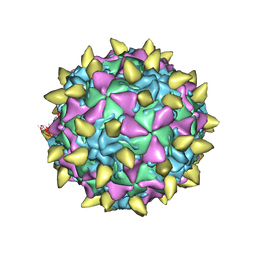 | | Complex of poliovirus with VHH PVSP19B | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBG
 
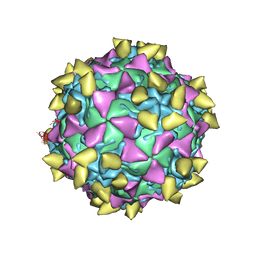 | | Complex of poliovirus with VHH PVSS21E | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
3JBI
 
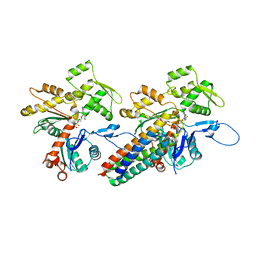 | | MDFF model of the vinculin tail domain bound to F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
3JBJ
 
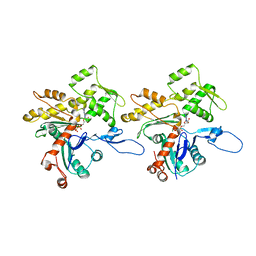 | | Cryo-EM reconstruction of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
3JBK
 
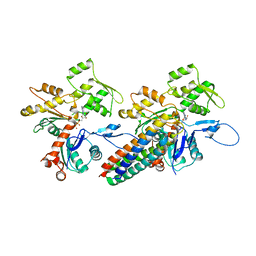 | | Cryo-EM reconstruction of the metavinculin-actin interface | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
3JBM
 
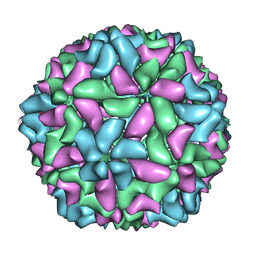 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
3JBN
 
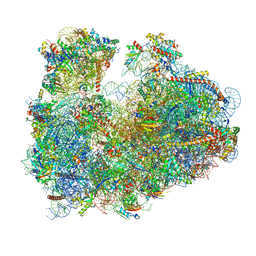 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JBO
 
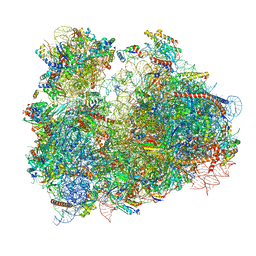 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P/E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JBP
 
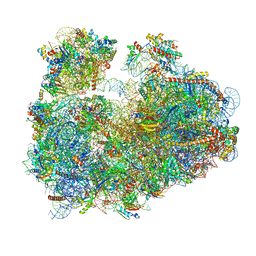 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JBQ
 
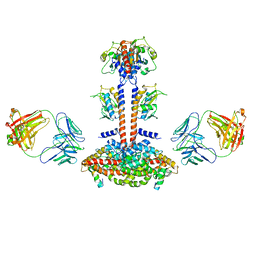 | | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6 | | Descriptor: | GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, IgG1-kappa 2E8 heavy chain, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
3JBR
 
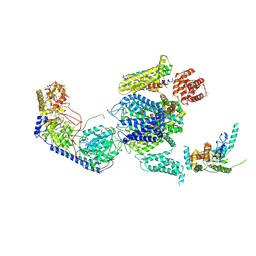 | | Cryo-EM structure of the rabbit voltage-gated calcium channel Cav1.1 complex at 4.2 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Voltage-dependent L-type calcium channel subunit alpha-1S, ... | | Authors: | Wu, J.P, Yan, Z, Yan, N. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 complex
Science, 350, 2015
|
|
3JBS
 
 | |
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
3JBU
 
 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | Deposit date: | 2015-10-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
3JBV
 
 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | Deposit date: | 2015-10-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
3JBW
 
 | | Cryo-electron microscopy structure of RAG Paired Complex (with NBD, no symmetry) | | Descriptor: | 12-RSS signal end forward strand, 5'-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', Nicked 12-RSS intermediate reverse strand, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3JBX
 
 | | Cryo-electron microscopy structure of RAG Signal End Complex (C2 symmetry) | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', 5'-D(*GP*CP*GP*AP*TP*GP*GP*TP*TP*AP*AP*CP*CP*A)-3', 5'-D(P*GP*TP*CP*TP*GP*TP*AP*GP*CP*AP*CP*TP*GP*TP*G)-3', ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
