3IW5
 
 | | Human p38 MAP Kinase in Complex with an Indole Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-(3-{[2-(2,3-dihydro-1,4-benzodioxin-6-ylamino)-2-oxoethyl]sulfanyl}-1H-indol-1-yl)ethyl]-3-(trifluoromethyl)benzamide, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3IW6
 
 | | Human p38 MAP Kinase in Complex with a Benzylpiperazin-Pyrrol | | Descriptor: | Mitogen-activated protein kinase 14, ethyl 4-[(4-benzylpiperazin-1-yl)carbonyl]-1-ethyl-3,5-dimethyl-1H-pyrrole-2-carboxylate, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3IW7
 
 | | Human p38 MAP Kinase in Complex with an Imidazo-pyridine | | Descriptor: | 2-({4-[(4-benzylpiperidin-1-yl)carbonyl]benzyl}sulfanyl)-3H-imidazo[4,5-c]pyridine, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3IW8
 
 | | Structure of Inactive Human p38 MAP Kinase in Complex with a Thiazole-Urea | | Descriptor: | 1-{4-[(1S)-1-amino-2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}-3-(3-chloro-4-fluorophenyl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3IWA
 
 | | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris | | Descriptor: | CALCIUM ION, FAD-dependent pyridine nucleotide-disulphide oxidoreductase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris
To be Published
|
|
3IWB
 
 | | T. maritima AdoMetDC in processed form | | Descriptor: | S-adenosylmethionine decarboxylase | | Authors: | Bale, S, Kavita, B, Ealick, S.E. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Complexes of Thermotoga maritimaS-adenosylmethionine decarboxylase provide insights into substrate specificity.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3IWC
 
 | |
3IWD
 
 | |
3IWE
 
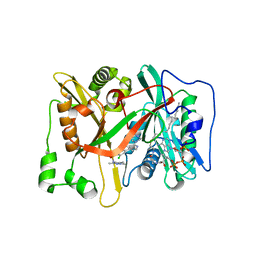 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646 | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646
To be Published
|
|
3IWF
 
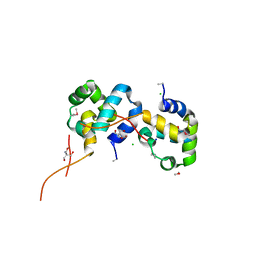 | | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Stein, A.J, Sather, A, Borovilos, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A
To be Published
|
|
3IWG
 
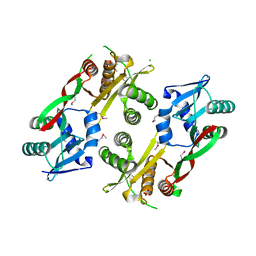 | | Acetyltransferase from GNAT family from Colwellia psychrerythraea. | | Descriptor: | Acetyltransferase, GNAT family, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Stein, A, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of Acetyltransferase from GNAT family from Colwellia psychrerythraea.
To be Published
|
|
3IWH
 
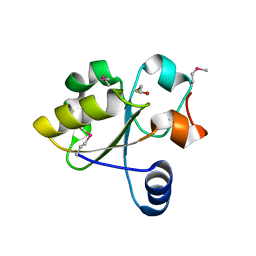 | | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus | | Descriptor: | BETA-MERCAPTOETHANOL, Rhodanese-like domain protein | | Authors: | Kim, Y, Chruszcz, M, Minor, W, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus
To be Published
|
|
3IWI
 
 | |
3IWJ
 
 | | Crystal structure of aminoaldehyde dehydrogenase 2 from Pisum sativum (PsAMADH2) | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative aminoaldehyde dehydrogenase, ... | | Authors: | Kopecny, D, Morera, S, Briozzo, P. | | Deposit date: | 2009-09-02 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional characterization of plant aminoaldehyde dehydrogenase from Pisum sativum with a broad specificity for natural and synthetic aminoaldehydes.
J.Mol.Biol., 396, 2010
|
|
3IWK
 
 | | Crystal structure of aminoaldehyde dehydrogenase 1 from Pisum sativum (PsAMADH1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Aminoaldehyde dehydrogenase, GLYCEROL, ... | | Authors: | Kopecny, D, Morera, S, Briozzo, P. | | Deposit date: | 2009-09-02 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of plant aminoaldehyde dehydrogenase from Pisum sativum with a broad specificity for natural and synthetic aminoaldehydes.
J.Mol.Biol., 396, 2010
|
|
3IWL
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cisplatin bound to a human copper chaperone.
J.Am.Chem.Soc., 131, 2009
|
|
3IWM
 
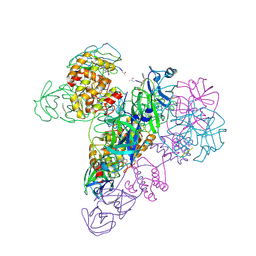 | | The octameric SARS-CoV main protease | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | Deposit date: | 2009-09-02 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
3IWN
 
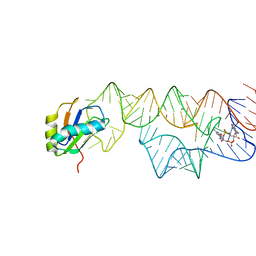 | | Co-crystal structure of a bacterial c-di-GMP riboswitch | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C-di-GMP riboswitch, U1 small nuclear ribonucleoprotein A | | Authors: | Kulshina, N, Baird, N.J, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3IWO
 
 | |
3IWP
 
 | |
3IWQ
 
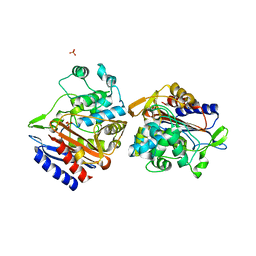 | |
3IWR
 
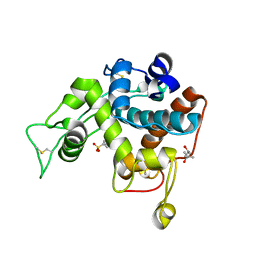 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chitinase | | Authors: | Kezuka, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2009-09-03 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
3IWT
 
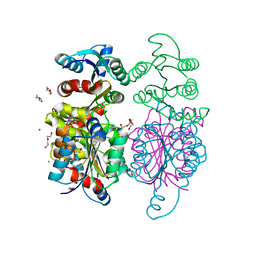 | | Structure of hypothetical molybdenum cofactor biosynthesis protein B from Sulfolobus tokodaii | | Descriptor: | 178aa long hypothetical molybdenum cofactor biosynthesis protein B, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3IWU
 
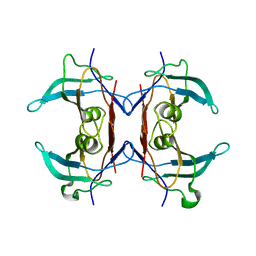 | | Crystal structure of Y116T/I16A double mutant of 5-hydroxyisourate hydrolase | | Descriptor: | 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Berni, R, Percudani, R, Zanotti, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
3IWV
 
 | | Crystal structure of Y116T mutant of 5-HYDROXYISOURATE HYDROLASE (TRP) | | Descriptor: | 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Berni, R, Percudani, R, Zanotti, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
