6PJA
 
 | |
6PJ5
 
 | |
6PJ9
 
 | |
8S0M
 
 | |
8S0L
 
 | |
8S0N
 
 | |
6P14
 
 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form B) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, ACETATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93001127 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6P68
 
 | |
5K5E
 
 | | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | De la Mora, E, Dighe, S.N, Deora, G.S, Ross, B.P, Nachon, F, Brazzolotto, X. | | Deposit date: | 2016-05-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening.
J.Med.Chem., 59, 2016
|
|
6PGI
 
 | | Asymmetric functions of a binuclear metal cluster within the transport pathway of the ZIP transition metal transporters | | Descriptor: | BbZIP, CADMIUM ION | | Authors: | Zhang, T, Sui, D, Zhang, C, Logan, T, Hu, J. | | Deposit date: | 2019-06-24 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetric functions of a binuclear metal center within the transport pathway of a human zinc transporter ZIP4.
Faseb J., 34, 2020
|
|
8SLB
 
 | | X-ray structure of CorA N-terminal domain in complex with conformation-specific synthetic antibody C12 | | Descriptor: | CHLORIDE ION, Cobalt/magnesium transport protein CorA, sAB C12 Heavy Chain, ... | | Authors: | Dominik, P.K, Erramilli, S.K, Reddy, B.G, Kossiakoff, A.A. | | Deposit date: | 2023-04-21 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Conformation-specific Synthetic Antibodies Discriminate Multiple Functional States of the Ion Channel CorA.
J.Mol.Biol., 435, 2023
|
|
6PJ7
 
 | |
6P13
 
 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form A) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6PJQ
 
 | |
6P95
 
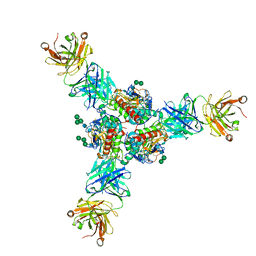 | | Structure of Lassa virus glycoprotein in complex with Fab 25.6A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Hastie, K.M. | | Deposit date: | 2019-06-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Convergent Structures Illuminate Features for Germline Antibody Binding and Pan-Lassa Virus Neutralization.
Cell, 178, 2019
|
|
8TAZ
 
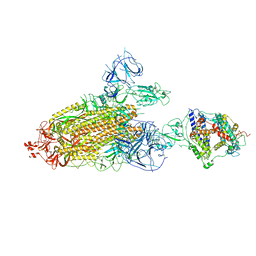 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Liang, B. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
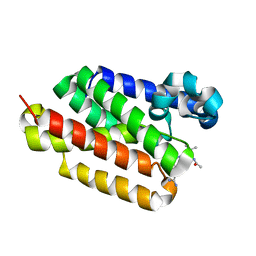
6PJ8
 
 | |
6PJP
 
 | |
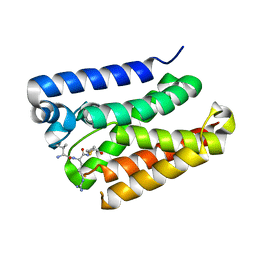
6PJ4
 
 | |
5JUL
 
 | | Near atomic structure of the Dark apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | Authors: | Cheng, T.C, Akey, I.V, Yuan, S, Yu, Z, Ludtke, S.J, Akey, C.W. | | Deposit date: | 2016-05-10 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A Near-Atomic Structure of the Dark Apoptosome Provides Insight into Assembly and Activation.
Structure, 25, 2017
|
|
8TEO
 
 | | Shaker in low K+ (4mM K+) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-07-06 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Eukaryotic Kv channel Shaker inactivates through selectivity filter dilation rather than collapse.
Sci Adv, 9, 2023
|
|
6PWF
 
 | |
6QF7
 
 | | Crystal structures of the recombinant beta-Factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, ... | | Authors: | Pathak, M, Mannal, R, Li, C, Bubacarr, G.K, Badraldin, K.H, Belviso, B.D, Camila, R.B, Dreveny, I, Fischer, P.M, Dekker, L.V, Oliva, M.L.V, Emsley, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structures of the recombinant beta-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8PWV
 
 | |
8Q5D
 
 | |
