3V12
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
8B5G
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-3-methyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7,8-dimethoxy-3-methyl-1~{H}-3-benzazepin-2-one, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
3T73
 
 | | Thermolysin In Complex With UBTLN22 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Water makes the difference: rearrangement of water solvation layer triggers non-additivity of functional group contributions in protein-ligand binding.
Chemmedchem, 7, 2012
|
|
1PRO
 
 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-98881 | | Descriptor: | (5R,6R)-2,4-BIS-(4-HYDROXY-3-METHOXYBENZYL)-1,5-DIBENZYL-3-OXO-6-HYDROXY-1,2,4-TRIAZACYCLOHEPTANE, HIV-1 PROTEASE | | Authors: | Park, C.H, Kong, X.P, Dealwis, C.G. | | Deposit date: | 1995-07-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel, picomolar inhibitor of human immunodeficiency virus type 1 protease.
J.Med.Chem., 39, 1996
|
|
1Y09
 
 | |
1Y8W
 
 | | T-To-T(High) quaternary transitions in human hemoglobin: alphaR92A oxy (2mM IHP, 20% PEG) (10 test sets) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, OXYGEN MOLECULE, ... | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2004-12-13 | | Release date: | 2005-01-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic evidence for a new ensemble of ligand-induced allosteric transitions in hemoglobin: the T-to-T(high) quaternary transitions.
Biochemistry, 44, 2005
|
|
6C16
 
 | |
3UNQ
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4Q9L
 
 | | P-glycoprotein cocrystallised with QZ-Phe | | Descriptor: | (30F)F(30F)F(30F)F Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1LZO
 
 | | Plasmodium Falciparum Triosephosphate Isomerase-Phosphoglycolate Complex | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate Isomerase | | Authors: | Parthasarathy, S, Balaram, H, Balaram, P, Murthy, M.R. | | Deposit date: | 2002-06-11 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Plasmodium falciparum triosephosphate isomerase-phosphoglycolate complex in two crystal forms: characterization of catalytic loop open and closed conformations in the ligand-bound state
Biochemistry, 41, 2002
|
|
4JFT
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor N-desmethyl-4-epi-(+)-Codonopsinine | | Descriptor: | (2S,3S,4R,5S)-2-(4-methoxyphenyl)-5-methylpyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
7GSB
 
 | |
4JFS
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor 4-epi-(+)-Codonopsinine | | Descriptor: | (2S,3S,4R,5S)-2-(4-methoxyphenyl)-1,5-dimethylpyrrolidine-3,4-diol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
1LYX
 
 | | Plasmodium Falciparum Triosephosphate Isomerase (PfTIM)-Phosphoglycolate complex | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate Isomerase | | Authors: | Parthasarathy, S, Balaram, H, Balaram, P, Murthy, M.R. | | Deposit date: | 2002-06-10 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Plasmodium falciparum triosephosphate isomerase-phosphoglycolate complex in two crystal forms: characterization of catalytic loop open and closed conformations in the ligand-bound state
Biochemistry, 41, 2002
|
|
4RUG
 
 | |
3PM3
 
 | | Bovine trypsin variant X(tripleIle227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2010-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4JFU
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor | | Descriptor: | (2S,3R,4S,5S)-2-methyl-5-(4-methylphenyl)pyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
3P2V
 
 | | Novel Benzothiazepine Inhibitor in Complex with human Aldose Reductase | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(2S)-4-oxo-2-phenyl-3,4-dihydro-1,5-benzothiazepin-5(2H)-yl]acetic acid | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-10-04 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
4OS5
 
 | |
2Y4N
 
 | | PaaK1 in complex with phenylacetyl adenylate | | Descriptor: | 2-PHENYLACETIC ACID, 5'-O-[HYDROXY(PHENYLACETYL)PHOSPHORYL]ADENOSINE, BETA-MERCAPTOETHANOL, ... | | Authors: | Law, A, Boulanger, M.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Defining a Structural and Kinetic Rationale for Paralogous Copies of Phenylacetate-Coa Ligases from the Cystic Fibrosis Pathogen Burkholderia Cenocepacia J2315.
J.Biol.Chem., 286, 2011
|
|
6HVK
 
 | | Pepducin UT-Pep2 a biased allosteric agonist of Urotensin-II receptor | | Descriptor: | Urotensin-2 receptor | | Authors: | Carotenuto, A, Hoang, T.A, Nassour, H, Martin, R.D, Billard, E, Myriam, L, Novellino, E, Tanny, J.C, Fournier, A, Hebert, T.E, Chatenet, D. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lipidated peptides derived from intracellular loops 2 and 3 of the urotensin II receptor act as biased allosteric ligands.
J.Biol.Chem., 297, 2021
|
|
6LCW
 
 | | Crosslinked alpha(Ni)-beta(Ni) human hemoglobin A in the T quaternary structure at 95 K: Dark | | Descriptor: | BUT-2-ENEDIAL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3QAS
 
 | | Structure of Undecaprenyl Diphosphate synthase | | Descriptor: | Undecaprenyl pyrophosphate synthase | | Authors: | Cao, R, Oldfield, E. | | Deposit date: | 2011-01-11 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Applying Molecular Dynamics Simulations to Identify Rarely Sampled Ligand-bound Conformational States of Undecaprenyl Pyrophosphate Synthase, an Antibacterial Target.
Chem.Biol.Drug Des., 77, 2011
|
|
4OS6
 
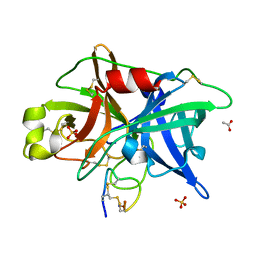 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK604 (bicyclic 2) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS1
 
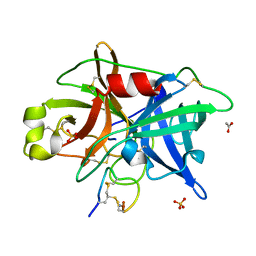 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK601 (bicyclic 1) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
