8QRT
 
 | |
8QS0
 
 | |
4Q27
 
 | |
3W9R
 
 | | Crystal structure of the high-affinity abscisic acid receptor PYL9/RCAR9 bound to ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9, HEXAETHYLENE GLYCOL | | Authors: | Nakagawa, M, Hirano, Y, Kagiyama, M, Shibata, N, Hakoshima, T. | | Deposit date: | 2013-04-13 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of high-affinity abscisic acid binding to PYL9/RCAR1.
Genes Cells, 19, 2014
|
|
3WNS
 
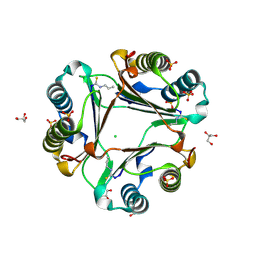 | | Allyl isothiocyanate inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Rutledge, M.T, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
3WQN
 
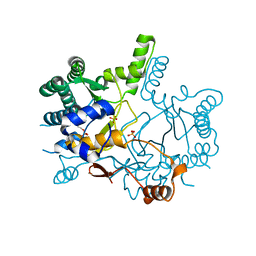 | | Crystal structure of Rv3378c_Y51F with TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3WG8
 
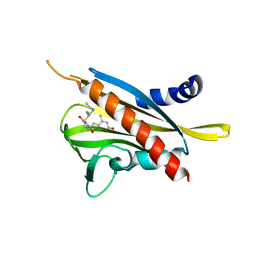 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist AS6 | | Descriptor: | (2Z,4E)-5-[(1S)-3-(hexylsulfanyl)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Okamoto, M, Muto, T, Endo, A, Nambara, E, Hirai, N, Ohnishi, T, Cutler, S.R, Todoroki, Y, Yajima, S. | | Deposit date: | 2013-07-31 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions
Nat.Chem.Biol., 10, 2014
|
|
3ZSX
 
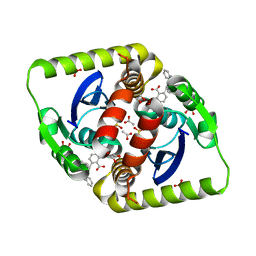 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-({[2-(benzylcarbamoyl)benzyl](prop-2-en-1-yl)amino}methyl)-1,3-benzodioxole-4-carboxylate, ACETATE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
4AFE
 
 | | Nek2 bound to hybrid compound 21 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-AMINO-5-{4-[(DIMETHYLAMINO)METHYL]THIOPHEN-2-YL}PYRIDIN-3-YL)-2-{[(1R,2Z)-4,4,4-TRIFLUORO-1-METHYLBUT-2-EN-1-YL]OXY}BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Yeoh, S, Innocenti, P, Hoelder, S, Bayliss, R. | | Deposit date: | 2012-01-19 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Design of Potent and Selective Hybrid Inhibitors of the Mitotic Kinase Nek2: Structure-Activity Relationship, Structural Biology, and Cellular Activity.
J.Med.Chem., 55, 2012
|
|
4AJ4
 
 | | rat LDHA in complex with 4-((2-allylsulfanyl-1,3-benzothizol-6-yl) amino)-4-oxo-butanoic acid | | Descriptor: | 4-oxo-4-{[2-(prop-2-en-1-ylsulfanyl)-1,3-benzothiazol-6-yl]amino}butanoic acid, L-LACTATE DEHYDROGENASE A CHAIN, MALONATE ION | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Vogtherr, M, Ward, R, Tart, J, Davies, G. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
4A9N
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-cyclopropyl-5-(3,5- dimethyl-1,2-oxazol-4-yl)-2-methylbenzene-1-sulfonamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN CONTAINING 2, DIMETHYL SULFOXIDE, ... | | Authors: | Chung, C, Bamborough, P. | | Deposit date: | 2011-11-26 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery of Bromodomain Inhibitors Part 2: Optimization of Phenylisoxazole Sulfonamides.
J.Med.Chem., 55, 2012
|
|
3ZSO
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based design | | Descriptor: | 5-{[(2-{[bis(4-methoxyphenyl)methyl]carbamoyl}benzyl)(prop-2-en-1-yl)amino]methyl}-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, INTEGRASE, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Deadman, J.J, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZOH
 
 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZSR
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (2S)-2-(2-carboxyethyl)-6-{[{2-[(cyclohexylmethyl)carbamoyl]benzyl}(prop-2-en-1-yl)amino]methyl}-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZVU
 
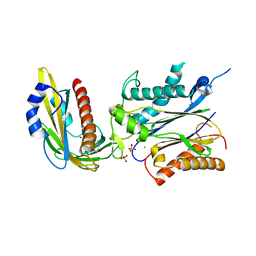 | | Structure of the PYR1 His60Pro mutant in complex with the HAB1 phosphatase and Abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABSCISIC ACID RECEPTOR PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thermodynamic Switch Modulates Abscisic Acid Receptor Sensitivity.
Embo J., 30, 2011
|
|
3ZK5
 
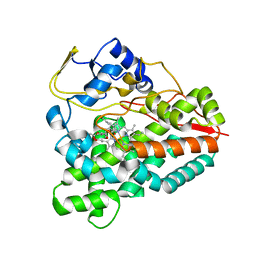 | | PikC D50N mutant bound to the 10-DML analog with the 3-(N,N-dimethylamino)ethanoate anchoring group | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ethyl-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl N,N-dimethylglycinate, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M. | | Deposit date: | 2013-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Directing Group-Controlled Regioselectivity in an Enzymatic C-H Bond Oxygenation.
J.Am.Chem.Soc., 136, 2014
|
|
4TRI
 
 | |
3ZOG
 
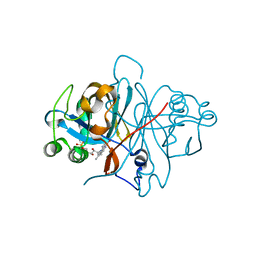 | | Crystal structure of FMN-binding protein (NP_142786.1) from Pyrococcus horikoshii with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
4CEZ
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-2-(2-carboxyethyl)-6-{[{2-[(cyclohexylmethyl)carbamoyl]benzyl}(prop-2-en-1-yl)amino]methyl}-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CE9
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-2-(2-carboxyethyl)-6-{[{2-[(cyclohexylmethyl)carbamoyl]benzyl}(prop-2-en-1-yl)amino]methyl}-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CRT
 
 | | Crystal structure of human monoamine oxidase B in complex with the multi-target inhibitor ASS234 | | Descriptor: | (E)-N-methyl-N-[[1-methyl-5-[3-[1-(phenylmethyl)piperidin-4-yl]propoxy]indol-2-yl]methyl]prop-1-en-1-amine, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Esteban, G, Allan, J, Samadi, A, Mattevi, A, Unzeta, M, Marco-Contelles, J, Binda, C, Ramsay, R.R. | | Deposit date: | 2014-02-28 | | Release date: | 2014-04-02 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Analysis of the Irreversible Inhibition of Human Monoamine Oxidases by Ass234, a Multi-Target Compound Designed for Use in Alzheimer'S Disease.
Biochim.Biophys.Acta, 1844, 2014
|
|
4Y9B
 
 | | Crystal structure of V30M mutated transthyretin in complex with alpha-mangostin | | Descriptor: | 1,3,6-trihydroxy-7-methoxy-2,8-bis(3-methylbut-2-en-1-yl)-9H-xanthen-9-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2015-02-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of gamma-Mangostin as an Amyloidogenesis Inhibitor
Sci Rep, 5, 2015
|
|
4Y9F
 
 | | Crystal structure of V30M mutated transthyretin with bromide in complex with gamma-mangostin | | Descriptor: | 1,3,6,7-tetrahydroxy-2,8-bis(3-methylbut-2-en-1-yl)-9H-xanthen-9-one, BROMIDE ION, Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2015-02-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of gamma-Mangostin as an Amyloidogenesis Inhibitor
Sci Rep, 5, 2015
|
|
4Y9E
 
 | | Crystal structure of V30M mutated transthyretin in complex with gamma-mangostin | | Descriptor: | 1,3,6,7-tetrahydroxy-2,8-bis(3-methylbut-2-en-1-yl)-9H-xanthen-9-one, CHLORIDE ION, Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2015-02-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of gamma-Mangostin as an Amyloidogenesis Inhibitor
Sci Rep, 5, 2015
|
|
6FDK
 
 | |
