4POL
 
 | | Crystal structures of thioredoxin with mesna at 2.8A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
8D7I
 
 | |
4WVP
 
 | | Crystal structure of an activity-based probe HNE complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BTN-3V3-NLB-OMT-OIC-3V2, ... | | Authors: | Lechtenberg, B.C, Kasperkiewicz, P, Robinson, H.R, Drag, M, Riedl, S.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Elastase-PK101 Structure: Mechanism of an Ultrasensitive Activity-based Probe Revealed.
Acs Chem.Biol., 10, 2015
|
|
8DYG
 
 | | IL17A homodimer bound to Compound 7 | | Descriptor: | (5P)-2-hydroxy-5-(6-methylquinolin-5-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYH
 
 | | IL17A homodimer bound to Compound 6 | | Descriptor: | (5P)-N-benzyl-6-chloro-5-(quinolin-5-yl)pyridin-3-amine, GLYCEROL, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYF
 
 | | IL17A homodimer bound to Compound 10 | | Descriptor: | (5M)-3-[({2-[2-(2-{2-[2-({[(5M)-3-carboxy-5-(5,8-dihydroquinolin-4-yl)phenyl]amino}methyl)phenoxy]ethoxy}ethoxy)ethoxy]phenyl}methyl)amino]-5-(quinolin-4-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYI
 
 | | IL17A homodimer bound to Compound 5 | | Descriptor: | (5P)-5-[5-(benzylamino)pyridin-3-yl]-N-[2-(morpholin-4-yl)ethyl]-1H-indazol-3-amine, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DY5
 
 | |
8DY1
 
 | |
8E3R
 
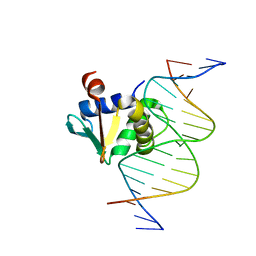 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*TP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
4OO5
 
 | |
1MDM
 
 | | INHIBITED FRAGMENT OF ETS-1 AND PAIRED DOMAIN OF PAX5 BOUND TO DNA | | Descriptor: | C-ETS-1 PROTEIN, PAIRED BOX PROTEIN PAX-5, PAX5/ETS BINDING SITE ON THE MB-1 PROMOTER | | Authors: | Garvie, C.W, Pufall, M.A, Graves, B.J, Wolberger, C. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STRUCTURAL ANALYSIS OF THE AUTOINHIBITION OF ETS-1 AND ITS ROLE IN PROTEIN PARTNERSHIPS
J.Biol.Chem., 277, 2002
|
|
5LGD
 
 | | The CIDRa domain from MCvar1 PfEMP1 bound to CD36 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hsieh, F.L, Higgins, M.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structural basis for CD36 binding by the malaria parasite.
Nat Commun, 7, 2016
|
|
4POK
 
 | | Crystal structures of thioredoxin with mesna at 2.5A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
4POM
 
 | | Crystal structures of thioredoxin with mesna at 1.85A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
2ERE
 
 | |
2ERG
 
 | |
2ER8
 
 | |
3X2W
 
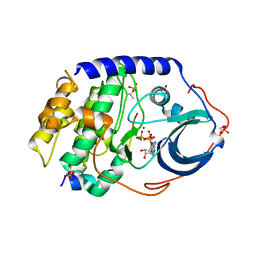 | | Michaelis complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
3KC0
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 10b | | Descriptor: | Fructose-1,6-bisphosphatase 1, [(8H-indeno[1,2-d][1,3]thiazol-4-yloxy)methyl]phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KBZ
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 6 | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(2-amino-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KC1
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 19a | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(7-carbamoyl-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6YCS
 
 | | Human Transcription Cofactor PC4 DNA-binding domain in complex with full phosphorothioate 5-10-5 2'-O-methyl DNA gapmer antisense oligonucleotide. | | Descriptor: | DNA (5'-D(P*(OKQ))-D(P*(OKT))-R(P*(RFJ))-D(*(OKQ)P*(OKT)P*(AS)P*(GS)P*(OKN)P*(OKN)P*(PST)P*(OKN)P*(PST)P*(GS)P*(GS)P*(AS)P*(OKT)P*(OKT))-3'), PC4 protein, SODIUM ION, ... | | Authors: | Hyjek-Skladanowska, M, Vickers, T.A, Napiorkowska, A, Anderson, B, Tanowitz, M, Crooke, S.T, Liang, X, Seth, P.P, Nowotny, M. | | Deposit date: | 2020-03-19 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Origins of the Increased Affinity of Phosphorothioate-Modified Therapeutic Nucleic Acids for Proteins.
J.Am.Chem.Soc., 142, 2020
|
|
5JR7
 
 | | Crystal structure of an ACRDYS heterodimer [RIa(92-365):C] of PKA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Bruystens, J.G.H, Wu, J, Taylor, S.S. | | Deposit date: | 2016-05-05 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structure of a PKA RI alpha Recurrent Acrodysostosis Mutant Explains Defective cAMP-Dependent Activation.
J. Mol. Biol., 428, 2016
|
|
7EZP
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
