5TBB
 
 | |
1LXK
 
 | | Streptococcus pneumoniae Hyaluronate Lyase in Complex with Tetrasaccharide Hyaluronan Substrate | | Descriptor: | Hyaluronate Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jedrzejas, M.J, Mello, L.V, De Groot, B.L, Li, S. | | Deposit date: | 2002-06-05 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate.
J.Biol.Chem., 277, 2002
|
|
3GCU
 
 | | Human P38 MAP kinase in complex with RL48 | | Descriptor: | 1-{3-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 14, ... | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-02-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors.
J.Am.Chem.Soc., 131, 2009
|
|
2YSA
 
 | | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1) | | Descriptor: | Retinoblastoma-binding protein 6, ZINC ION | | Authors: | Ohnishi, S, Sato, M, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1)
To be Published
|
|
5O3H
 
 | | Human Brd2(BD2) mutant in complex with 9-ME-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-9-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-propanamide, (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
3KKG
 
 | |
2BU5
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | 4-({(2R,5S)-2,5-DIMETHYL-4-[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]PIPERAZIN-1-YL}CARBONYL)BENZONITRILE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
7BO8
 
 | | A hexameric de novo coiled-coil assembly: CC-Type2-(VaYd)4-Y3F-W19(BrPhe)-Y24F. | | Descriptor: | 1,2-ETHANEDIOL, CC-Type2-(VaYd)4-Y3F-W19(BrPhe)-Y24F, OXAMIC ACID | | Authors: | Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-05-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Coiled-Coil Assemblies Accommodate Multiple Aromatic Residues.
Biomacromolecules, 22, 2021
|
|
4R5Z
 
 | | Crystal structure of Rv3772 encoded aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
5TUG
 
 | |
5TRZ
 
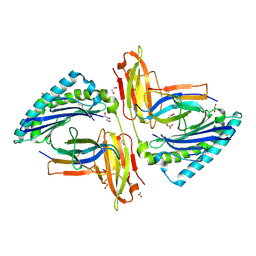 | | Crystal structure of MHC-I H2-KD complexed with peptides of Mycobacterial tuberculosis (YQSGLSIVM) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-10-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | MHC-restricted Ag85B-specific CD8+T cells are enhanced by recombinant BCG prime and DNA boost immunization in mice.
Eur.J.Immunol., 2019
|
|
4R6V
 
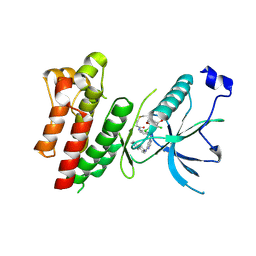 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex with FIIN-3, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3KMH
 
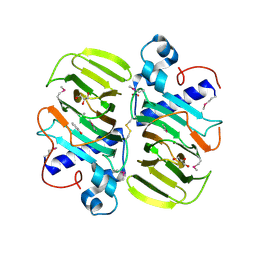 | |
5TS3
 
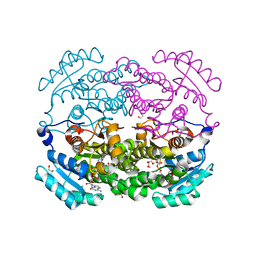 | |
2X74
 
 | | Human foamy virus integrase - catalytic core. | | Descriptor: | INTEGRASE | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Legrand, P, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3HYG
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (2R)-N~4~-hydroxy-2-(3-hydroxybenzyl)-N~1~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
2WLR
 
 | |
5TWS
 
 | | Post-catalytic complex of human Polymerase Mu (H329A) with newly incorporated UTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5TZF
 
 | | Structure of the BldD CTD(D116A)-(c-di-GMP)2 intermediate, form 1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
4RE5
 
 | | Acylaminoacyl peptidase complexed with a chloromethylketone inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Acylamino-acid-releasing enzyme, ... | | Authors: | Menyhard, D.K, Orgovan, Z, Szeltner, Z, Szamosi, I, Harmat, V. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytically distinct states captured in a crystal lattice: the substrate-bound and scavenger states of acylaminoacyl peptidase and their implications for functionality.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5TY8
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-phenyl-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
6MK4
 
 | |
4D98
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in space group H32 at pH 7.5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Santos, C.R, Meza, A.N, Martins, N.H, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2012-01-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
5U0D
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-(2,4-dimethoxyphenyl)-2-sulfanylidene-1,3-oxazolidin-4-one, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
