3UN6
 
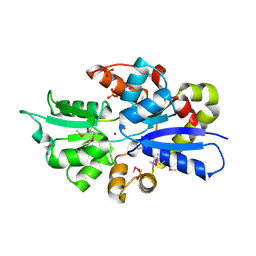 | | 2.0 Angstrom Crystal Structure of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Zinc bound | | Descriptor: | ABC transporter substrate-binding protein, PHOSPHATE ION, ZINC ION | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-15 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2.0 Angstrom Crystal Structure of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Zinc bound.
TO BE PUBLISHED
|
|
3UPD
 
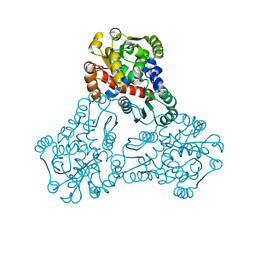 | | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus.
TO BE PUBLISHED
|
|
4KTV
 
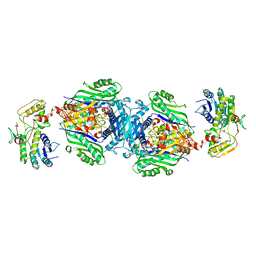 | | Structural insights of MAT enzymes: MATa2b complexed with adenosine and pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, MAGNESIUM ION, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-05-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
4K9F
 
 | | Neutron structure of Perdeuterated Rubredoxin refined against 1.75 resolution data collected on the new IMAGINE instrument at HFIR, ORNL | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-04-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | The IMAGINE instrument: first neutron protein structure and new capabilities for neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3U80
 
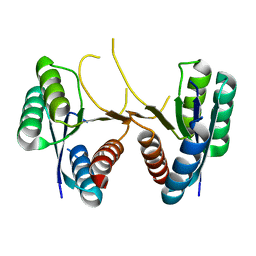 | | 1.60 Angstrom Resolution Crystal Structure of a 3-Dehydroquinate Dehydratase-like Protein from Bifidobacterium longum | | Descriptor: | 3-dehydroquinate dehydratase, type II | | Authors: | Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a type II dehydroquinate dehydratase-like protein from Bifidobacterium longum.
J.Struct.Funct.Genom., 14, 2013
|
|
3TX1
 
 | | X-ray crystal structure of Listeria monocytogenes EGD-e UDP-N-acetylenolpyruvylglucosamine reductase (MurB) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | X-ray crystal structure of Listeria monocytogenes EGD-e UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
TO BE PUBLISHED
|
|
3U2E
 
 | | EAL domain of phosphodiesterase PdeA in complex with 5'-pGpG and Mg++ | | Descriptor: | GGDEF family protein, MAGNESIUM ION, RNA (5'-R(P*GP*G)-3') | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | EAL domain from Caulobacter crescentus in complex with 5'-pGpG and Mg++
To be Published
|
|
3S6W
 
 | | Crystal structure of Tudor domain of human TDRD3 | | Descriptor: | ISOPROPYL ALCOHOL, Tudor domain-containing protein 3 | | Authors: | Liu, H.P, Xu, R.M. | | Deposit date: | 2011-05-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30
Plos One, 7, 2012
|
|
3RXS
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamine (F04 and A06, cocktail experiment) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3SBE
 
 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3S5C
 
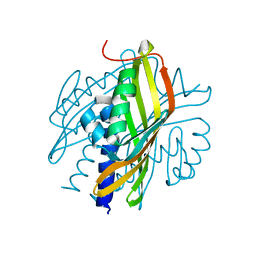 | | Crystal Structure of a Hexachlorocyclohexane dehydrochlorinase (LinA) Type2 | | Descriptor: | LinA | | Authors: | Kukshal, V, Macwan, A.S, Kumar, A, Ramachandran, R. | | Deposit date: | 2011-05-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the hexachlorocyclohexane dehydrochlorinase (LinA-type2): mutational analysis, thermostability and enantioselectivity
Plos One, 7, 2012
|
|
3SAI
 
 | |
3SBD
 
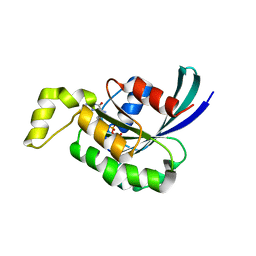 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
3S6T
 
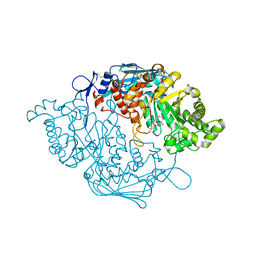 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 V327G complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Yang, Q, Shen, X. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases.
Biochem.J., 438, 2011
|
|
3SA1
 
 | |
3SHU
 
 | | Crystal structure of ZO-1 PDZ3 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
3SJ7
 
 | |
9FEM
 
 | |
9FER
 
 | |
3S7K
 
 | | Structure of thrombin mutant Y225P in the E form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, POTASSIUM ION, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
3SH9
 
 | | Crystal structure of fluorophore-labeled beta-lactamase PenP in complex with cefotaxime | | Descriptor: | 1-[6-(dimethylamino)naphthalen-2-yl]ethanone, Beta-lactamase, CEFOTAXIME, ... | | Authors: | Wong, W.-T, Zhao, Y.-X, Leung, Y.-C. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increased structural flexibility at the active site of a fluorophore-conjugated beta-lactamase distinctively impacts its binding toward diverse cephalosporin antibiotics
J.Biol.Chem., 286, 2011
|
|
3SA2
 
 | |
3SGI
 
 | |
2XME
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE, GLYCEROL | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Archaeoglobus Fulgidus Ctp:Inositol-1-Phosphate Cytidylyltransferase, a Key Enzyme for Di-Myo-Inositol-Phosphate Synthesis in (Hyper)Thermophiles.
J.Bacteriol., 193, 2011
|
|
6JG2
 
 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-nitrophenoxy)-3,5-bis(oxidanyl)oxan-4-yl]sulfanyl-oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
