3K9O
 
 | | The crystal structure of E2-25K and UBB+1 complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of E2-25K/UBB+1 Interaction for Neurotoxicity of Alzheimer Disease by Proteasome Inhibition
To be Published
|
|
5A01
 
 | | O-GlcNAc transferase from Drososphila melanogaster | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, O-GLYCOSYLTRANSFERASE | | Authors: | Mariappa, D, Zheng, X, Schimpl, M, Raimi, O, Rafie, K, Ferenbach, A.T, Mueller, H.J, van Aalten, D.M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Dual functionality of O-GlcNAc transferase is required for Drosophila development.
Open Biol, 5, 2015
|
|
5TYZ
 
 | | DNA Polymerase Mu Product Complex, Mn2+ (960 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
12E8
 
 | | 2E8 FAB FRAGMENT | | Descriptor: | IGG1-KAPPA 2E8 FAB (HEAVY CHAIN), IGG1-KAPPA 2E8 FAB (LIGHT CHAIN) | | Authors: | Rupp, B, Trakhanov, S. | | Deposit date: | 1998-03-14 | | Release date: | 1998-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a monoclonal 2E8 Fab antibody fragment specific for the low-density lipoprotein-receptor binding region of apolipoprotein E refined at 1.9 A.
Acta Crystallogr.,Sect.D, null, 1999
|
|
5U0G
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-[(2,4-dimethoxyphenyl)methyl]-1,3-thiazolidine-2,4-dione, 1,2-ETHANEDIOL, Carbonic anhydrase 2, ... | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5TZO
 
 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
5U0E
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-benzyl-2-sulfanylidene-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, Carbonic anhydrase 2, ... | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
7FN2
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06G08 from the F2X-Universal Library | | Descriptor: | 1-[2-(morpholin-4-yl)pyridin-4-yl]methanamine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
6BGC
 
 | | The crystal structure of the W145A variant of TpMglB-2 (Tp0684) with bound glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/galactose-binding lipoprotein, ... | | Authors: | Brautigam, C.A, Norgard, M.V, Deka, R.K. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Crystal structures of MglB-2 (TP0684), a topologically variant d-glucose-binding protein from Treponema pallidum, reveal a ligand-induced conformational change.
Protein Sci., 27, 2018
|
|
5A6Q
 
 | | Native structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-06-30 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
7FNL
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07D01 from the F2X-Universal Library | | Descriptor: | 1-ethyl-N-[(thiophen-2-yl)methyl]-1H-tetrazol-5-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5TR4
 
 | | Structure of Ubiquitin activating enzyme (Uba1) in complex with ubiquitin and TAK-243 | | Descriptor: | Ubiquitin, Ubiquitin-activating enzyme E1 1, [(1~{R},2~{R},3~{S},4~{R})-2,3-bis(oxidanyl)-4-[[2-[3-(trifluoromethylsulfanyl)phenyl]pyrazolo[1,5-a]pyrimidin-7-yl]amino]cyclopentyl]methyl sulfamate | | Authors: | Sintchak, M.D. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and Characterization of a Small Molecule Inhibitor of the Ubiquitin Activating Enzyme (TAK-243)
To be published
|
|
5TRK
 
 | |
3JTK
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055 | | Descriptor: | (2R)-3-benzyl-2-(2-bromo-4-hydroxy-5-methoxyphenyl)-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055
To be Published
|
|
6B25
 
 | |
7V1A
 
 | | Stapled TBS peptide from RIAM bound to talin R7R8 domains | | Descriptor: | 1,2-ETHANEDIOL, ASP-ILE-ASP-GLN-MET-PHE-SER-THR-LEU-LEU-GLY-GLU-MK8-ASP-LEU-LEU-MK8-GLN-SER, Talin-1 | | Authors: | Zhang, P, Gao, T, Wu, J. | | Deposit date: | 2022-05-11 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibition of talin-induced integrin activation by a double-hit stapled peptide.
Structure, 31, 2023
|
|
5WZU
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 24 | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[[2-(trifluoromethyl)phenyl]methyl]indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZW
 
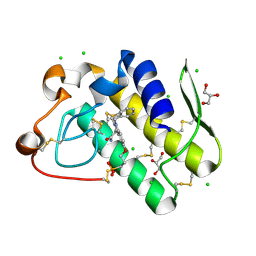 | | Crystal structure of human secreted phospholipase A2 group IIE with LY311727 | | Descriptor: | (3-{[3-(2-amino-2-oxoethyl)-1-benzyl-2-ethyl-1H-indol-5-yl]oxy}propyl)phosphonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
8X9Q
 
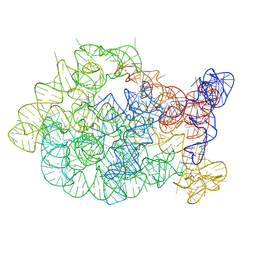 | | Cte-A695U-branching,Mg,active | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (809-MER) | | Authors: | Ling, X.B, Ma, J.B. | | Deposit date: | 2023-11-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of a natural circularly permuted group II intron reveal mechanisms of branching and backsplicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5UBI
 
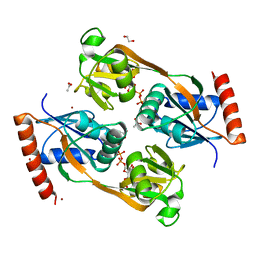 | | Catalytic core domain of Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni with bound PRPP | | Descriptor: | 1,2-ETHANEDIOL, 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Mittelstaedt, G, Jiao, W, Livingstone, E.K, Parker, E.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase.
Biochem. J., 475, 2018
|
|
5AKK
 
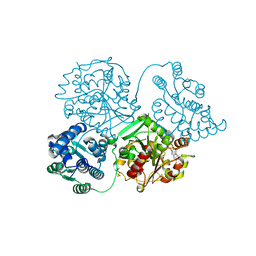 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 6-bromo-1,3-dihydro-2H-indol-2-one, BIFUNCTIONAL EPOXIDE HYDROLASE 2, SULFATE ION | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
4O5J
 
 | | Crystal structure of SabA from Helicobacter pylori | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Uncharacterized protein | | Authors: | Pang, S.S, Nguyen, S.T.S, Whisstock, J.C. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori
J.Biol.Chem., 289, 2013
|
|
7FPJ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P10D05 from the F2X-Universal Library | | Descriptor: | 1-(1-benzyl-1H-imidazol-2-yl)methanamine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5TYC
 
 | | DNA Polymerase Mu Reactant Complex, 10mM Mg2+ (15 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
2AJX
 
 | |
