4EZ4
 
 | | free KDM6B structure | | Descriptor: | Lysine-specific demethylase 6B, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Cheng, Z.J, Patel, D.J. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response.
Nature, 488, 2012
|
|
4GAZ
 
 | | Crystal Structure of a Jumonji Domain-containing Protein JMJD5 | | Descriptor: | Lysine-specific demethylase 8, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, H, Zhou, X, Zhang, X, Tao, Y, Chen, N, Zang, J. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of a Jumonji Domain-containing Protein JMJD5
To be Published
|
|
6RK9
 
 | |
5UHO
 
 | |
5UHL
 
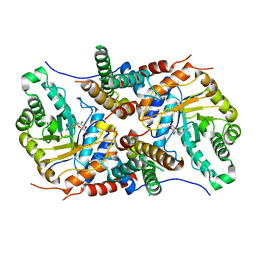 | | Crystal structure of the core catalytic domain of human O-GlcNAcase complexed with Thiamet G | | Descriptor: | (2Z,3aR,5R,6S,7R,7aR)-2-(ethylimino)-5-(hydroxymethyl)hexahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Elsen, N.L. | | Deposit date: | 2017-01-11 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the core catalytic domain of human O-GlcNAcase and molecular basis of activity and inhibition
To Be Published
|
|
6HKI
 
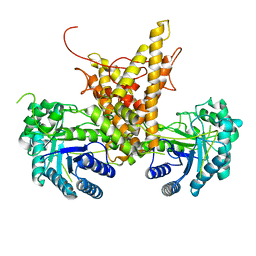 | |
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5JQY
 
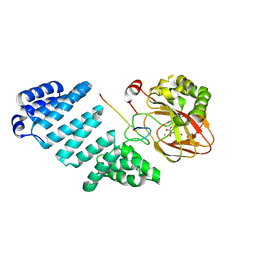 | |
5JZA
 
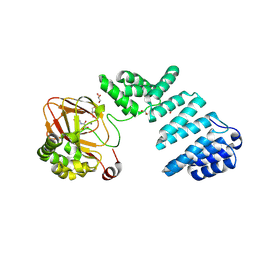 | |
5JZU
 
 | |
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5JZ8
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH)oxygenase and TPR domains in complex with manganese, N-oxalylglycine, and factor X substrate peptide fragment (39mer) | | Descriptor: | ACETATE ION, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | McDonough, M.A, Pfeffer, I. | | Deposit date: | 2016-05-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Aspartate/asparagine-beta-hydroxylase crystal structures reveal an unexpected epidermal growth factor-like domain substrate disulfide pattern.
Nat Commun, 10, 2019
|
|
6F4T
 
 | | Human JMJD5 (W414C) in complex with Mn(II), NOG and RCCD1 (139-143) (complex-5) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, JmjC domain-containing protein 5, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
6F0W
 
 | | prolyl hydroxylase in complex with hypoxia inducible factor oxygen degradation domain peptide fragment from Trichoplax adhaerens | | Descriptor: | HIF prolyl hydroxylase, Hypoxia inducible factor, alpha subunit, ... | | Authors: | McDonough, M.A, Boleininger, A, Schofield, C.J. | | Deposit date: | 2017-11-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Born to sense: biophysical analyses of the oxygen sensing prolyl hydroxylase from the simplest animal Trichoplax adhaerens.
Hypoxia (Auckl), 6, 2018
|
|
6F4R
 
 | | Human JMJD5 (N308C) in complex with Mn(II), NOG and RCCD1 (139-143) (complex-3) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, JmjC domain-containing protein 5, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
5L9V
 
 | |
5LAS
 
 | |
5LA9
 
 | |
5L9R
 
 | |
2OQ6
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OQ7
 
 | | The crystal structure of JMJD2A complexed with Ni and N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
4QKF
 
 | | Crystal structure of human ALKBH7 in complex with N-oxalylglycine and Mn(II) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 7, mitochondrial, MANGANESE (II) ION, ... | | Authors: | Wang, G, Chen, Z. | | Deposit date: | 2014-06-06 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism
J.Biol.Chem., 289, 2014
|
|
2OS2
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys36 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OT7
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide monomethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-07 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
6H8P
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Histone H1.4(18-32)K26me3 peptide (15-mer) | | Descriptor: | CHLORIDE ION, GLYCEROL, Histone H1.4, ... | | Authors: | Chowdhury, R, Walport, L.J, Schofield, C.J. | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26.
FEBS Lett., 592, 2018
|
|
