2PJV
 
 | |
159L
 
 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
6UXP
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylglycerol | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), Bcl-2 homologous antagonist/killer, GLYCEROL | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3EMQ
 
 | | Crystal structure of xilanase XynB from Paenibacillus barcelonensis complexed with an inhibitor | | Descriptor: | (1S,2S,3R,6R)-6-[(2-hydroxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | Authors: | Sanz-Aparicio, J, Isorna, P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
169L
 
 | |
5FJY
 
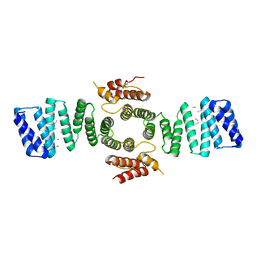 | | Crystal structure of mouse kinesin light chain 2 (residues 161-480) | | Descriptor: | KINESIN LIGHT CHAIN 2, UNKNOWN PEPTIDE | | Authors: | Pernigo, S, Yip, Y.Y, Sanger, A, Xu, M, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Light Chains of Kinesin-1 are Autoinhibited.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9FCI
 
 | | USP1 bound to KSQ-4279 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 6-(4-cyclopropyl-6-methoxy-pyrimidin-5-yl)-1-[[4-[1-propan-2-yl-4-(trifluoromethyl)imidazol-2-yl]phenyl]methyl]pyrazolo[3,4-d]pyrimidine, Polyubiquitin-C, Ubiquitin carboxyl-terminal hydrolase 1, ... | | Authors: | Rennie, M.L, Gundogdu, M, Walden, H. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279.
J.Med.Chem., 67, 2024
|
|
5L6H
 
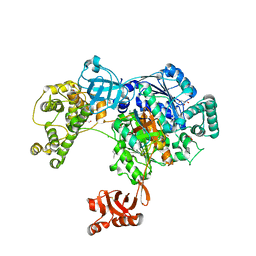 | | Uba1 in complex with Ub-ABPA3 covalent adduct | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
6V14
 
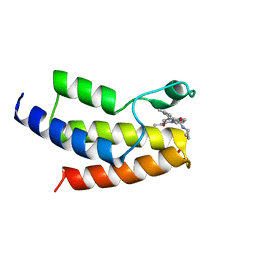 | | Crystal structure of the bromodomain of human BRD9 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 9 | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1D
 
 | | Crystal structure of human trefoil factor 1 | | Descriptor: | Trefoil factor 1 | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Scott, N.E, Goddard-Borger, E.D. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Trefoil factors share a lectin activity that defines their role in mucus.
Nat Commun, 11, 2020
|
|
6IIM
 
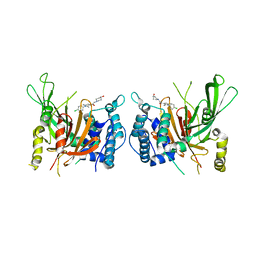 | | USP14 catalytic domain with IU1-206 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(4-hydroxypiperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6DNA
 
 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
5C1I
 
 | | m1A58 tRNA methyltransferase mutant - D170A | | Descriptor: | SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Ponchon, L, Degut, C, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-14 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
9FCJ
 
 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (ordered subset, focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Polyubiquitin-C, Ubiquitin carboxyl-terminal hydrolase 1, ... | | Authors: | Rennie, M.L, Gundogdu, M, Walden, H. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279.
J.Med.Chem., 67, 2024
|
|
7D24
 
 | | Hsp90 alpha N-terminal domain in complex with a 4B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
5YWX
 
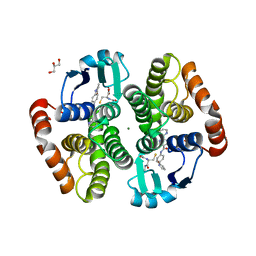 | | Crystal structure of hematopoietic prostaglandin D synthase in complex with F092 | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Omura, A, Tanaka, A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Characterization of crystal water molecules in a high-affinity inhibitor and hematopoietic prostaglandin D synthase complex by interaction energy studies.
Bioorg. Med. Chem., 26, 2018
|
|
6BSD
 
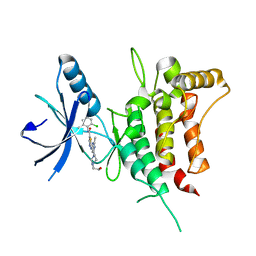 | | DDR1 bound to Dasatinib | | Descriptor: | Epithelial discoidin domain-containing receptor 1, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Georghiou, G, Seeliger, M.A. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | What Makes a Kinase Promiscuous for Inhibitors?
Cell Chem Biol, 26, 2019
|
|
2PVE
 
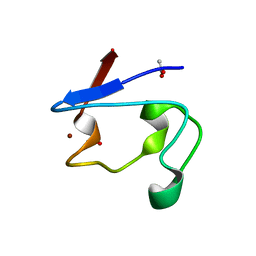 | | NMR and X-ray Analysis of Structural Additivity in Metal Binding Site-Swapped Hybrids of Rubredoxin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Rubredoxin, ... | | Authors: | LeMaster, D.M, Anderson, J.S, Wang, L, Guo, Y, Li, H, Hernandez, G. | | Deposit date: | 2007-05-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.79 Å) | | Cite: | NMR and X-ray analysis of structural additivity in metal binding site-swapped hybrids of rubredoxin.
Bmc Struct.Biol., 7, 2007
|
|
7D26
 
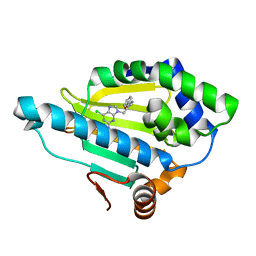 | | Hsp90 alpha N-terminal domain in complex with a 8 compund | | Descriptor: | 6-chloranyl-9-[(2-phenyl-1,3-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7H6G
 
 | | THE 1.21 A CRYSTAL STRUCTURE OF HUMAN CATHEPSIN G IN COMPLEX WITH N-[2-[6-fluoro-2-[(4-hydroxy-5-methyl-2-oxo-5-phenylfuran-3-yl)-phenylmethyl]-1H-indol-3-yl]ethyl]acetamide | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cathepsin G, N-(2-{6-fluoro-2-[(R)-[(5R)-4-hydroxy-5-methyl-2-oxo-5-phenyl-2,5-dihydrofuran-3-yl](phenyl)methyl]-1H-indol-3-yl}ethyl)acetamide, ... | | Authors: | Banner, D.W, Benz, J.M, Schlatter, D, Hilpert, H. | | Deposit date: | 2024-04-19 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structures of human Chymase and Cathepsin G
To be published
|
|
5FH1
 
 | | The structure of rat cytosolic PEPCK variant E89D in complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
7YRE
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(N137A,Q140S,Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
5CAL
 
 | | EGFR kinase domain mutant "TMLR" with compound 24 | | Descriptor: | 2,2-dimethyl-3-[(4-{[2-methyl-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-yl]amino}pyrimidin-2-yl)amino]propanamide, Epidermal growth factor receptor | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
6V1U
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to TP-472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
