4Z1O
 
 | |
7HK3
 
 | |
3N2K
 
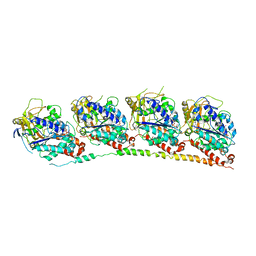 | | TUBULIN-NSC 613862: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Barbier, P, Dorleans, A, Devred, F, Sanz, L, Allegro, D, Alfonso, C, Knossow, M, Peyrot, V, Andreu, J.M. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers.
J.Biol.Chem., 285, 2010
|
|
7LAY
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD3 bound to SG3-179 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 3, ... | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
5TXT
 
 | | Structure of asymmetric apo/holo ALAS dimer from S. cerevisiae | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 5-aminolevulinate synthase, mitochondrial, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
4R06
 
 | | Crystal Structure of SR2067 bound to PPARgamma | | Descriptor: | 1-(naphthalen-1-ylsulfonyl)-N-[(1S)-1-phenylpropyl]-1H-indole-5-carboxamide, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Marrewijk, L, Kamenecka, T, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-07-30 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | SR2067 Reveals a Unique Kinetic and Structural Signature for PPAR gamma Partial Agonism.
Acs Chem.Biol., 11, 2016
|
|
6LKB
 
 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
5IYW
 
 | |
6FDX
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1086 | | Descriptor: | (4aS,8aR)-2-[1-(2-aminoquinazolin-4-yl)piperidin-4-yl]-4-(3,4-dimethoxyphenyl)-1,2,4a,5,8,8a-hexahydrophthalazin-1-one, FORMIC ACID, GUANIDINE, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | TbrPDEB1 structure with inhibitor NPD-1086
To be published
|
|
7HJD
 
 | |
7HJH
 
 | |
6SW6
 
 | |
4JQR
 
 | |
2V3Q
 
 | | Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, HUMAN PHOSPHATE BINDING PROTEIN, ... | | Authors: | Morales, R, Berna, A, Carpentier, P, Elias, M, Contreras-Martel, C, Renault, F, Nicodeme, M, Chesne-Seck, M.-L, Bernier, F, Dupuy, J, Schaeffer, C, Diemer, H, Van Dorsselaer, A, Fontecilla, J.C, Masson, P, Rochu, D, Chabriere, E. | | Deposit date: | 2007-06-20 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Tandem Use of X-Ray Crystallography and Mass Spectrometry to Obtain Ab Initio the Complete and Exact Amino Acids Sequence of Hpbp, a Human 38kDa Apolipoprotein
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
9EJH
 
 | | Peptide-independent T cell receptor recognition of HLA-DQ2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, G9 T cell receptor alpha chain, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A naturally selected alpha beta T cell receptor binds HLA-DQ2 molecules without co-contacting the presented peptide.
Nat Commun, 16, 2025
|
|
4OVY
 
 | | Crystal structure of Haloacid dehalogenase domain protein hydrolase from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Chang, C, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Haloacid dehalogenase domain protein hydrolase from Planctomyces limnophilus DSM 3776
To be published
|
|
1RBS
 
 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
6T1M
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
4OAD
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, GNAT superfamily acetyltransferase PA4794, ... | | Authors: | Majorek, K.A, Niedzialkowska, E, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with chloramphenicol
To be Published
|
|
9L8I
 
 | | Rhodothermus marines cellobiose 2-epimerase RmCE in complex with mannobiose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Saburi, W, Muto, H, Jaito, N, Kato, K, Yu, J, Yao, M, Mori, H. | | Deposit date: | 2024-12-27 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural analysis of the mechanism for the catalysis and specificity of cellobiose 2-epimerase from Rhodothermus marinus.
Biosci.Biotechnol.Biochem., 89, 2025
|
|
4ZAU
 
 | | AZD9291 complex with wild type EGFR | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Squire, C.J, Yosaatmadja, Y, Flanagan, J.U, McKeage, M. | | Deposit date: | 2015-04-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed.
J.Struct.Biol., 192, 2015
|
|
7EL9
 
 | | Structure of Machupo virus L polymerase in complex with Z protein and 3'-vRNA (dimeric complex) | | Descriptor: | MANGANESE (II) ION, Machupo virus 3'-vRNA promoter, RING finger protein Z, ... | | Authors: | Peng, R, Xu, X, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
9KWH
 
 | | Crystal structure of S. aureus tryptophanyl-tRNA synthetase complexed with chuangxinmycin | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, Tryptophan--tRNA ligase | | Authors: | Ren, Y, Qiao, H, Wang, S, Liu, W, Fang, P. | | Deposit date: | 2024-12-05 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic insights into the ATP-mediated and species-dependent inhibition of TrpRS by chuangxinmycin.
Rsc Chem Biol, 6, 2025
|
|
3I85
 
 | |
9KXB
 
 | | Crystal structure of S. aureus tryptophanyl-tRNA synthetase complexed with 3-methylchuangxinmycin | | Descriptor: | (6~{R})-5,5-dimethyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, Tryptophan--tRNA ligase | | Authors: | Ren, Y, Qiao, H, Wang, S, Liu, W, Fang, P. | | Deposit date: | 2024-12-06 | | Release date: | 2025-07-23 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanistic insights into the ATP-mediated and species-dependent inhibition of TrpRS by chuangxinmycin.
Rsc Chem Biol, 6, 2025
|
|
