3PXU
 
 | |
4HYV
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei in the presence of Magnesium, PEP and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2012-11-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | `In crystallo' substrate binding triggers major domain movements and reveals magnesium as a co-activator of Trypanosoma brucei pyruvate kinase.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HYW
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei in the presence of Magnesium and F26BP | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2012-11-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | `In crystallo' substrate binding triggers major domain movements and reveals magnesium as a co-activator of Trypanosoma brucei pyruvate kinase.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1HOZ
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX | | Descriptor: | CALCIUM ION, GLYCEROL, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
3SMS
 
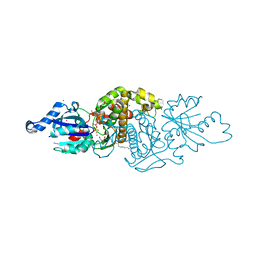 | | Human Pantothenate kinase 3 in complex with a pantothenate analog | | Descriptor: | (2R)-N-[3-(heptylamino)-3-oxopropyl]-2,4-dihydroxy-3,3-dimethylbutanamide, ADENOSINE-5'-DIPHOSPHATE, Pantothenate kinase 3, ... | | Authors: | Mottaghi, K, Tempel, W, Hong, B, Smil, D, Bolshan, Y, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human Pantothenate kinase 3 in complex with a pantothenate analog
to be published
|
|
5FSV
 
 | |
3F5M
 
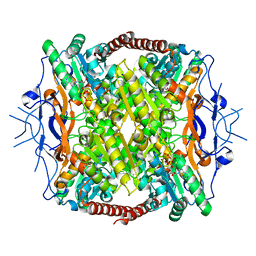 | | Crystal Structure of ATP-Bound Phosphofructokinase from Trypanosoma brucei | | Descriptor: | 6-phospho-1-fructokinase (ATP-dependent phosphofructokinase), ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | McNae, I.W, Martinez-Oyanedel, J, Keillor, J.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of ATP-bound phosphofructokinase from Trypanosoma brucei reveals conformational transitions different from those of other phosphofructokinases.
J.Mol.Biol., 385, 2009
|
|
5K4W
 
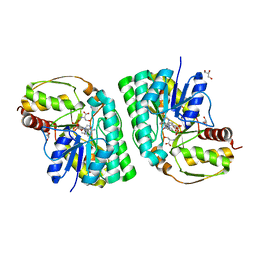 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NADH and L-threonine refined to 1.72 angstroms | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-threonine 3-dehydrogenase, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5K4U
 
 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei showing different active site loop conformations between dimer subunits, refined to 1.9 angstroms | | Descriptor: | ACETATE ION, GLYCEROL, L-threonine 3-dehydrogenase, ... | | Authors: | Adjogatse, E.K, Cooper, J.B, Erskine, P.T. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5GN5
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5K4Q
 
 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NAD+ refined to 2.3 angstroms | | Descriptor: | GLYCEROL, L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Adjogatse, E.K, Cooper, J.B, Erskine, P.T. | | Deposit date: | 2016-05-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2A0K
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.8 A resolution | | Descriptor: | GLYCEROL, Nucleoside 2-deoxyribosyltransferase, SULFATE ION | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
5E26
 
 | | Crystal structure of human PANK2: the catalytic core domain in complex with pantothenate and adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, LOPPNAU, P, RAVICHANDRAN, M, CHENG, C, TEMPEL, W, SEITOVA, A, HUTCHINSON, A, HONG, B.S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of human PANK2: the catalytic core domain in complex with pantothenate and adenosine diphosphate
to be published
|
|
6T98
 
 | | Trypanothione Reductase from Leishmania infantum in complex with 9a | | Descriptor: | 4-[3-methyl-1-[4-[4-(2-phenylethyl)-1,3-thiazol-2-yl]-3-(2-piperidin-4-ylethoxy)phenyl]-1,2,3-triazol-3-ium-4-yl]butan-1-amine, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
5GN6
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17b | | Descriptor: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5FSX
 
 | |
6T95
 
 | | Trypanothione Reductase from Leismania infantum in complex with 4a | | Descriptor: | 1-[2-[5-[4-(4-azanylbutyl)-3-methyl-1,2,3-triazol-3-ium-1-yl]-2-[4-(2-phenylethyl)-1,3-thiazol-2-yl]phenoxy]ethyl]imidazolidin-2-one, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
5FUV
 
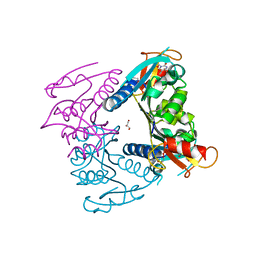 | | catalytic domain of Thymidine kinase from Trypanosoma brucei with dThd | | Descriptor: | GLYCEROL, PHOSPHATE ION, THYMDINE KINASE, ... | | Authors: | Timm, J, Valente, M, Castillo-Acosta, V, Balzarini, T, Nettleship, J.E, Rada, H, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2016-01-31 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cell Cycle Regulation and Novel Structural Features of Thymidine Kinase, an Essential Enzyme in Trypanosoma Brucei.
Mol.Microbiol., 102, 2016
|
|
6I7N
 
 | |
4NEV
 
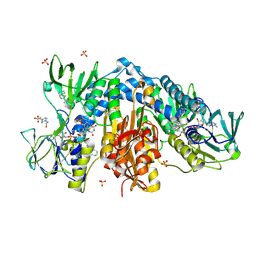 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
4NEW
 
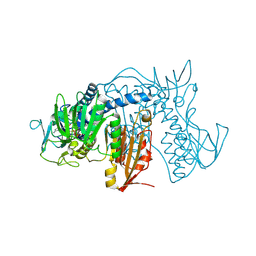 | | Crystal structure of Trypanothione Reductase from Trypanosoma cruzi in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, Trypanothione reductase, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
3UY4
 
 | | Crystal Structure of Pantoate--Beta-Alanine Ligase from Campylobacter jejuni complexed with AMP and vitamin B5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal Structure of Pantoate--Beta-Alanine Ligase from Campylobacter jejuni complexed with AMP and vitamin B5
To be Published
|
|
3UNV
 
 | | Pantoea agglomerans Phenylalanine Aminomutase | | Descriptor: | (3S)-3-amino-3-phenylpropanoic acid, 1,2-ETHANEDIOL, AdmH, ... | | Authors: | Geiger, J, Strom, S. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into the mechanistic pathway of the Pantoea agglomerans phenylalanine aminomutase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6SU2
 
 | |
6SPT
 
 | | High resolution crystal structure of N-terminal domain of PEX14 from Trypanosoma brucei in complex with the fist compound with sub-micromolar trypanocidal activity | | Descriptor: | 5-[(4-methoxynaphthalen-1-yl)methyl]-1-[2-[(2-methyl-1-oxidanyl-propan-2-yl)amino]ethyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Napolitano, V, Dawidowski, M, Kalel, V.C, Fino, R, Emmanouilidis, L, Lenhart, D, Ostertag, M, Kaiser, M, Kolonko, M, Schilebs, W, Maser, P, Tetko, I, Hadian, K, Plettenburg, O, Erdmann, R, Sattler, M, Popowicz, G.M, Dubin, G. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Activity Relationship in Pyrazolo[4,3-c]pyridines, First Inhibitors of PEX14-PEX5 Protein-Protein Interaction with Trypanocidal Activity.
J.Med.Chem., 63, 2020
|
|
