3GTM
 
 | | Co-complex of Backtracked RNA polymerase II with TFIIS | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTQ
 
 | | Backtracked RNA polymerase II complex induced by damage | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*CP*AP*TP*AP*AP*CP*CP*AP*CP*AP*GP*GP*CP*TP*CP*CP*TP*CP*TP*CP*CP*AP*TP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTL
 
 | | Backtracked RNA polymerase II complex with 13mer with G<>U mismatch | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTP
 
 | | Backtracked RNA polymerase II complex with 24mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
8EJ3
 
 | |
4YFN
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YFK
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YFX
 
 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
8RAP
 
 | |
8RAM
 
 | |
7B7U
 
 | | Cryo-EM structure of mammalian RNA polymerase II in complex with human RPAP2 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Dienemann, C, Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of mammalian RNA polymerase II in complex with human RPAP2.
Commun Biol, 4, 2021
|
|
1EF4
 
 | | SOLUTION STRUCTURE OF THE ESSENTIAL RNA POLYMERASE SUBUNIT RPB10 FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, ZINC ION | | Authors: | Mackereth, C.D, Arrowsmith, C.H, Edwards, A.M, Mcintosh, L.P, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-07 | | Release date: | 2000-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Zinc-bundle structure of the essential RNA polymerase subunit RPB10 from Methanobacterium thermoautotrophicum.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4RY6
 
 | | C-terminal mutant (W550A) of HCV/J4 RNA polymerase | | Descriptor: | HCV J4 RNA polymerase (NS5B) | | Authors: | Jaeger, J, Cherry, A, Dennis, C. | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
4RY7
 
 | | C-terminal mutant (D559E) of HCV/J4 RNA polymerase | | Descriptor: | HCV J4 RNA polymerase (NS5B) | | Authors: | Jaeger, J, Cherry, A, Dennis, C. | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
4RY5
 
 | | C-terminal mutant (W550N) of HCV/J4 RNA polymerase | | Descriptor: | HCV J4 RNA polymerase (NS5B), MANGANESE (II) ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Jaeger, J, Cherry, A, Dennis, C. | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
4RY4
 
 | | C-terminal mutant (Y448F) of HCV/J4 RNA polymerase | | Descriptor: | HCV J4 RNA polymerase (NS5B) | | Authors: | Jaeger, J, Cherry, A, Dennis, C. | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
2JA5
 
 | | CPD lesion containing RNA Polymerase II elongation complex A | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TTP*TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II
Science, 315, 2007
|
|
2JA8
 
 | | CPD lesion containing RNA Polymerase II elongation complex D | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TP*TTP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*AP*UP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
2JA7
 
 | | CPD lesion containing RNA Polymerase II elongation complex C | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TTP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
2JA6
 
 | | CPD lesion containing RNA Polymerase II elongation complex B | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TTP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | CPD damage recognition by transcribing RNA polymerase II.
Science, 315, 2007
|
|
6VJS
 
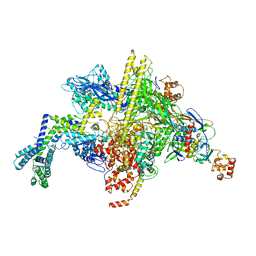 | | Escherichia coli RNA polymerase and ureidothiophene-2-carboxylic acid complex | | Descriptor: | 3-{[benzyl(ethyl)carbamoyl]amino}-5-(4-phenoxyphenyl)thiophene-2-carboxylic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murakami, K.S, Molodtsov, V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.02 Å) | | Cite: | Evaluation of Bacterial RNA Polymerase Inhibitors in a Staphylococcus aureus -Based Wound Infection Model in SKH1 Mice.
Acs Infect Dis., 6, 2020
|
|
6VR4
 
 | |
1ZYR
 
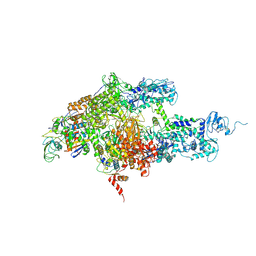 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
8EMB
 
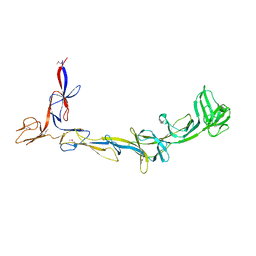 | | X-ray crystal structure of Thermosynechococcus elongatus Si3 domain of RNA polymerase RpoC2 subunit | | Descriptor: | DNA-directed RNA polymerase subunit beta' | | Authors: | Murakami, K.S, Imashimizu, M, Qayyum, M.Z, Vishwakarma, R.K, Yuzenkova, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F3C
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (38-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
