8UEL
 
 | | Crystal structure of enolase from Litopenaeus vannamei | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Chang, X, Zhao, G. | | Deposit date: | 2023-10-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and Structural Analyses of Enolase from Shrimp ( Litopenaeus vannamei ).
J.Agric.Food Chem., 71, 2023
|
|
6TV2
 
 | | Heme d1 biosynthesis associated Protein NirF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Protein NirF, ... | | Authors: | Kluenemann, T, Layer, G, Blankenfeldt, W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Crystal structure of NirF: insights into its role in heme d 1 biosynthesis.
Febs J., 288, 2021
|
|
7U9K
 
 | | Staphylococcus aureus D-alanine-D-alanine ligase in complex with ATP, D-ala-D-ala, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design and synthesis of ATP-competitive N-acyl-substituted sulfamide d-alanine-d-alanine ligase inhibitors.
Bioorg.Med.Chem., 96, 2023
|
|
5Z5O
 
 | | Structure of Pycnonodysostosis disease related I249T mutant of human cathepsin K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Biswas, S, Roy, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Not all pycnodysostosis-related mutants of human cathepsin K are inactive - crystal structure and biochemical studies of an active mutant I249T.
FEBS J., 285, 2018
|
|
4ZQS
 
 | | New compact conformation of linear Ub2 structure | | Descriptor: | ubiquitin | | Authors: | Thach, T.T, Shin, D, Han, S, Lee, S. | | Deposit date: | 2015-05-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7DFQ
 
 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7D1D
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutaminyl cyclase bound to 1-benzylimidazole | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, Glutamine cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2B
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Ni ion bound to the active site | | Descriptor: | Glutaminyl-peptide cyclotransferase, NICKEL (II) ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2I
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Fe ion bound to the active site | | Descriptor: | FE (III) ION, Glutaminyl-peptide cyclotransferase, SULFATE ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D23
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with one K ion bound to the active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutaminyl-peptide cyclotransferase, POTASSIUM ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1N
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with three Cu ions bound to the active site | | Descriptor: | BICARBONATE ION, COPPER (II) ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2J
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Cd ion bound to the active site | | Descriptor: | BICARBONATE ION, CADMIUM ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
6T6N
 
 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) in complex with NADH at 2.5 A resolution | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-[acyl-carrier protein] reductase, D-MALATE, ... | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
7PTZ
 
 | | High resolution X-ray structure of E. coli expressed Lentinus similis LPMO. | | Descriptor: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION | | Authors: | Banerjee, S, Muderspach, S.J, Tandrup, T, Ipsen, J.O, Hernandez-Rollan, C, Norholm, H.H.M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-09-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.093 Å) | | Cite: | Protonation State of an Important Histidine from High Resolution Structures of Lytic Polysaccharide Monooxygenases.
Biomolecules, 12, 2022
|
|
1PS3
 
 | | Golgi alpha-mannosidase II in complex with kifunensine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase II, ... | | Authors: | Shah, N, Kuntz, D.A, Rose, D.R. | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of Kifunensine and 1-Deoxymannojirimycin Binding to Class I and II alpha-Mannosidases Demonstrates Different Saccharide Distortions in Inverting and Retaining Catalytic Mechanisms
Biochemistry, 42, 2003
|
|
6TYR
 
 | |
4H2H
 
 | | Crystal structure of an enolase (mandalate racemase subgroup, target EFI-502101) from Pelagibaca bermudensis htcc2601, with bound mg and l-4-hydroxyproline betaine (betonicine) | | Descriptor: | (2S,4R)-4-hydroxy-1,1-dimethylpyrrolidinium-2-carboxylate, (4S)-2-METHYL-2,4-PENTANEDIOL, IODIDE ION, ... | | Authors: | Vetting, M.W, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
6TV9
 
 | |
6FHP
 
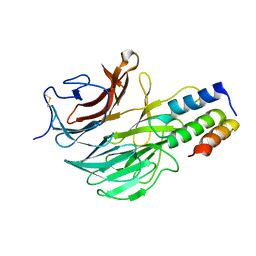 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
5YGS
 
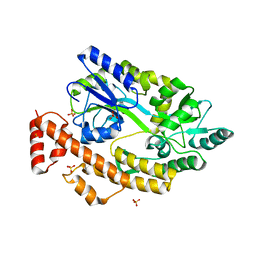 | | Human TNFRSF25 death domain | | Descriptor: | Human TNRSF25 death domain, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-26 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
6UBQ
 
 | |
4IWN
 
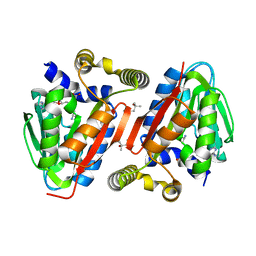 | | Crystal structure of a putative methyltransferase CmoA in complex with a novel SAM derivative | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, tRNA (cmo5U34)-methyltransferase | | Authors: | Aller, P, Lobley, C.M, Byrne, R.T, Antson, A.A, Waterman, D.G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6UCW
 
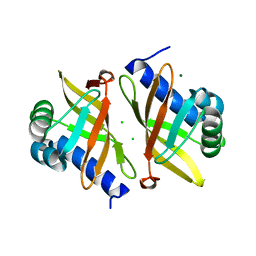 | | Multi-conformer model of Apo Ketosteroid Isomerase from Pseudomonas Putida (pKSI) at 250 K | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Steroid Delta-isomerase | | Authors: | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TZD
 
 | |
6U1Z
 
 | |
