1EMJ
 
 | | URACIL-DNA GLYCOSYLASE BOUND TO DNA CONTAINING A 4'-THIO-2'DEOXYURIDINE ANALOG PRODUCT | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(ASU)P*AP*TP*CP*TP*T)-3'), URACIL, ... | | Authors: | Parikh, S.S, Walcher, G, Jones, G.D, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2OXM
 
 | | Crystal structure of a UNG2/modified DNA complex that represent a stabilized short-lived extrahelical state in ezymatic DNA base flipping | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*(4MF)P*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*TP*CP*TP*T)-3'), Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
2OYT
 
 | | Crystal Structure of UNG2/DNA(TM) | | Descriptor: | DNA strand1, DNA strand2, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
1Q3F
 
 | | Uracil DNA glycosylase bound to a cationic 1-aza-2'-deoxyribose-containing DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*(NRI)P*AP*TP*CP*TP*T)-3', PHOSPHATE ION, ... | | Authors: | Bianchet, M.A, Seiple, L.A, Jiang, Y.L, Ichikawa, Y, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2003-07-29 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electrostatic guidance of glycosyl cation migration along the reaction coordinate of uracil DNA glycosylase.
Biochemistry, 42, 2003
|
|
1FLZ
 
 | | URACIL DNA GLYCOSYLASE WITH UAAP | | Descriptor: | URACIL, URACIL-DNA GLYCOSYLASE | | Authors: | Werner, R.M, Jiang, Y.L, Gordley, R.G, Jagadeesh, G.J, Ladner, J.E, Xiao, G, Tordova, M, Gilliland, G.L, Stivers, J.T. | | Deposit date: | 2000-08-15 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase.
Biochemistry, 39, 2000
|
|
2RBA
 
 | | Structure of Human Thymine DNA Glycosylase Bound to Abasic and Undamaged DNA | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DCP*DTP*DCP*DTP*DGP*DTP*DAP*DCP*DGP*DTP*DGP*DAP*DGP*DCP*DAP*DGP*DTP*DGP*DGP*DA)-3'), DNA (5'-D(*DCP*DCP*DAP*DCP*DTP*DGP*DCP*DTP*DCP*DAP*(3DR)P*DGP*DTP*DAP*DCP*DAP*DGP*DAP*DGP*DCP*DTP*DGP*DT)-3'), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Maiti, A, Pozharski, E, Drohat, A.C. | | Deposit date: | 2007-09-18 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2SSP
 
 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
1SSP
 
 | | WILD-TYPE URACIL-DNA GLYCOSYLASE BOUND TO URACIL-CONTAINING DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*(D1P)P*AP*TP*CP*TP*T)-3', URACIL, ... | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
6AIL
 
 | | CRYSTAL STRUCTURE AT 1.3 ANGSTROMS RESOLUTION OF A NOVEL UDG, UdgX, FROM Mycobacterium smegmatis | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase X | | Authors: | Ahn, W.C, Aroli, S, Varshney, V, Woo, E.J. | | Deposit date: | 2018-08-24 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.335 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJR
 
 | | Complex form of Uracil DNA glycosylase X and uracil | | Descriptor: | IRON/SULFUR CLUSTER, URACIL, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.341 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJO
 
 | | Complex form of Uracil DNA glycosylase X and uracil-DNA. | | Descriptor: | DNA (5'-D(P*(ORP)P*TP*T)-3'), IRON/SULFUR CLUSTER, PHOSPHATE ION, ... | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJP
 
 | | Complex form of Uracil DNA glycosylase X and deoxyuridine monophosphate. | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJQ
 
 | | E52Q mutant form of Uracil DNA glycosylase X from Mycobacterium smegmatis. | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.342 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJS
 
 | | H109S mutant form of Uracil DNA glycosylase X. | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
3CXM
 
 | |
3EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | Descriptor: | GLYCEROL, PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-10-13 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
3FCK
 
 | | Complex of UNG2 and a fragment-based design inhibitor | | Descriptor: | 3-({[3-({[(1E)-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methylidene]amino}oxy)propyl]amino}methyl)benzoic acid, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3FCI
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{(E)-[(3-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}propoxy)imino]methyl}benzoic acid, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3FCF
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-[(1E,7E)-8-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)-3,6-dioxa-2,7-diazaocta-1,7-dien-1-yl]benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3FCL
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{[(4-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}butyl)amino]methyl}benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
5EUG
 
 | | CRYSTALLOGRAPHIC AND ENZYMATIC STUDIES OF AN ACTIVE SITE VARIANT H187Q OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE: CRYSTAL STRUCTURES OF MUTANT H187Q AND ITS URACIL COMPLEX | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
5FF8
 
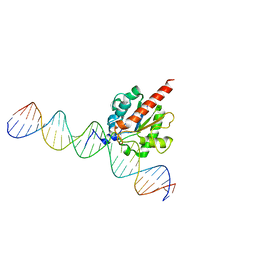 | | TDG enzyme-product complex | | Descriptor: | DNA, G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
5H0K
 
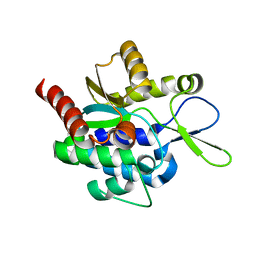 | |
5H0J
 
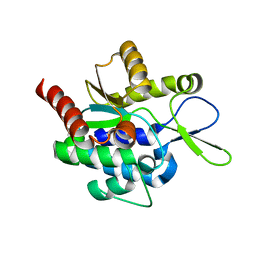 | |
5HF7
 
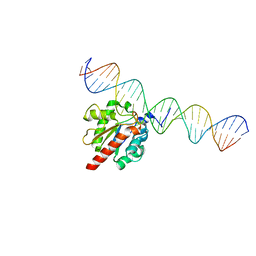 | | TDG enzyme-substrate complex | | Descriptor: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2016-01-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
