2ZM6
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kaminishi, T, Wang, H, Kawazoe, M, Ishii, R, Schluenzen, F, Hanawa-Suetsugu, K, Wilson, D.N, Nomura, M, Takemoto, C, Shirouzu, M, Fucini, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-11 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the Thermus thermophilus 30S ribosomal subunit
To be Published
|
|
2YKR
 
 | | 30S ribosomal subunit with RsgA bound in the presence of GMPPNP | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Guo, Q, Yuan, Y, Xu, Y, Feng, B, Liu, L, Chen, K, Lei, J, Gao, N. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural Basis for the Function of a Small Gtpase Rsga on the 30S Ribosomal Subunit Maturation Revealed by Cryoelectron Microscopy.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5OT7
 
 | | Elongation factor G-ribosome complex captures in the absence of inhibitors. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mace, K, Giudice, E, Chat, S, Gillet, R. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-14 | | Last modified: | 2018-04-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structure of an elongation factor G-ribosome complex captured in the absence of inhibitors.
Nucleic Acids Res., 46, 2018
|
|
1HNW
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH TETRACYCLINE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HR0
 
 | | CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Morgan-Warren, R.J, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-20 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an initiation factor bound to the 30S ribosomal subunit.
Science, 291, 2001
|
|
1HNX
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH PACTAMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HNZ
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1FKA
 
 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
1FJG
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
1V9F
 
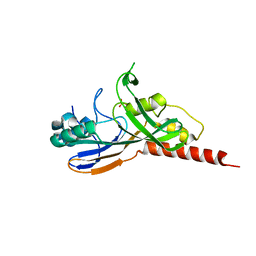 | | Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli | | Descriptor: | PHOSPHATE ION, Ribosomal large subunit pseudouridine synthase D | | Authors: | Mizutani, K, Machida, Y, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
5T2A
 
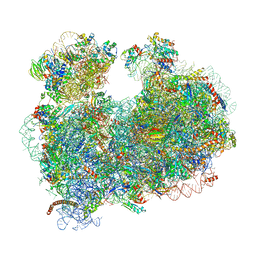 | | CryoEM structure of the Leishmania donovani 80S ribosome at 2.9 Angstrom resolution | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5TGA
 
 | |
5T2C
 
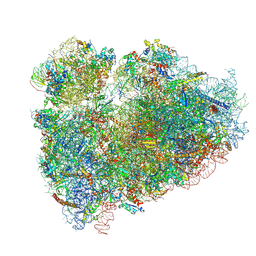 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
1VIO
 
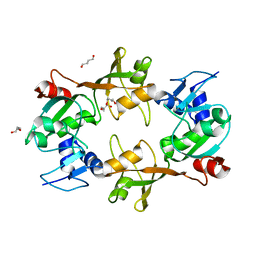 | | Crystal structure of pseudouridylate synthase | | Descriptor: | 1,4-BUTANEDIOL, Ribosomal small subunit pseudouridine synthase A | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the pseudouridine synthase RsuA from Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VVJ
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 bound to Codon CCC-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-05-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.440001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1VY6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing short substrate-mimic Cytidine-Puromycin in the A site and acylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6ERI
 
 | | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10 alpha, ... | | Authors: | Perez Borema, A, Aibara, S, Paul, B, Tobiasson, V, Kimanius, D, Forsberg, B.O, Wallden, K, Lindahl, E, Amunts, A. | | Deposit date: | 2017-10-18 | | Release date: | 2018-04-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor.
Nat Plants, 4, 2018
|
|
6ENJ
 
 | |
6ENU
 
 | | Polyproline-stalled ribosome in the presence of elongation-factor P (EF-P) | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Arenz, S, Wilson, D.N. | | Deposit date: | 2017-10-06 | | Release date: | 2017-11-22 | | Last modified: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.
Mol. Cell, 68, 2017
|
|
1N34
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit in the presence of codon and crystallographically disordered near-cognate transfer rna anticodon stem-loop mismatched at the first codon position | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Murphy IV, F.V, Tarry, M.J, Ramakrishnan, V. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form
Cell(Cambridge,Mass.), 111, 2002
|
|
6EML
 
 | | Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Heuer, A, Thomson, E, Schmidt, C, Berninghausen, O, Becker, T, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a late pre-40S ribosomal subunit fromSaccharomyces cerevisiae.
Elife, 6, 2017
|
|
4OX9
 
 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmA bound to the 30S ribosomal subunit | | Descriptor: | 16S rRNA, 16S rRNA (adenine(1408)-N(1))-methyltransferase, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Conn, G.L, Dunham, C.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8035 Å) | | Cite: | Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6ENF
 
 | | Cryo-EM structure of a polyproline-stalled ribosome in the absence of EF-P | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Arenz, S, Wilson, D.N. | | Deposit date: | 2017-10-04 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.
Mol. Cell, 68, 2017
|
|
4P70
 
 | | Crystal Structure of Unmodified tRNA Proline (CGG) Bound to Codon CCG on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2014-03-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
