8D1R
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT520 | | Descriptor: | 1-{3-[(4-chlorophenyl)methoxy]phenyl}methanamine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of substituted benzyloxy-benzylamine inhibitors of acetyltransferase Eis and their anti-mycobacterial activity.
Eur.J.Med.Chem., 242, 2022
|
|
8D25
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT530 | | Descriptor: | 1-{2-[(3-chlorophenyl)methoxy]phenyl}-N-[(pyridin-4-yl)methyl]methanamine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of substituted benzyloxy-benzylamine inhibitors of acetyltransferase Eis and their anti-mycobacterial activity.
Eur.J.Med.Chem., 242, 2022
|
|
8DA2
 
 | | Acinetobacter baumannii L,D-transpeptidase | | Descriptor: | L,D-transpeptidase family protein | | Authors: | Toth, M, Stewart, N.K, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2022-06-12 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The l,d-Transpeptidase Ldt Ab from Acinetobacter baumannii Is Poorly Inhibited by Carbapenems and Has a Unique Structural Architecture.
Acs Infect Dis., 8, 2022
|
|
7PRI
 
 | | Carbonic Anhydrase from Schistosoma Mansoni in complex with clorsulon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-azanyl-6-[1,2,2-tris(chloranyl)ethenyl]benzene-1,3-disulfonamide, Carbonic anhydrase, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-09-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Inhibition of Schistosoma mansoni carbonic anhydrase by the antiparasitic drug clorsulon: X-ray crystallographic and in vitro studies.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PG6
 
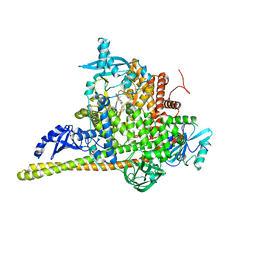 | | Crystal Structure of PI3Kalpha in complex with the inhibitor NVP-BYL719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49943733 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
6Y8D
 
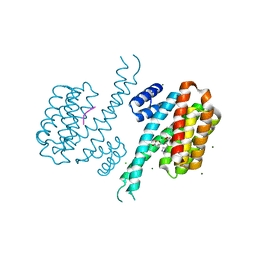 | | 14-3-3 Sigma in complex with phosphorylated caspase{pS164} peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6Y54
 
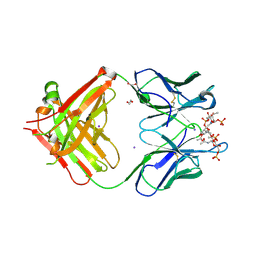 | | Crystal structure of a Neisseria meningitidis serogroup A capsular oligosaccharide bound to a functional Fab | | Descriptor: | 1,2-ETHANEDIOL, Fab A1.1 H chain, Fab A1.1 L chain, ... | | Authors: | Dello Iacono, L, Henriques, P, Adamo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure of a protective epitope reveals the importance of acetylation of Neisseria meningitidis serogroup A capsular polysaccharide.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y0R
 
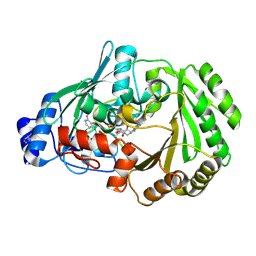 | | Chitooligosaccharide oxidase | | Descriptor: | Chitooligosaccharide oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Savino, S, Fraaije, M.W. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Analysis of the structure and substrate scope of chitooligosaccharide oxidase reveals high affinity for C2-modified glucosamines.
Febs Lett., 594, 2020
|
|
6Y8A
 
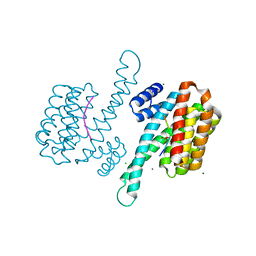 | | 14-3-3 Sigma in complex with phosphorylated camkk2{pS511} peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6Y8X
 
 | |
6YAO
 
 | | Crystal structure of ZmCKO4a in complex with inhibitor 1-[2-(2-Hydroxy-ethyl)-phenyl]-3-(3-trifluoromethoxy-phenyl)-urea | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(2-Hydroxy-ethyl)-phenyl]-3-(3-trifluoromethoxy-phenyl)-urea, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diphenylurea-derived cytokinin oxidase/dehydrogenase inhibitors for biotechnology and agriculture.
J.Exp.Bot., 72, 2021
|
|
6YC5
 
 | |
8DAX
 
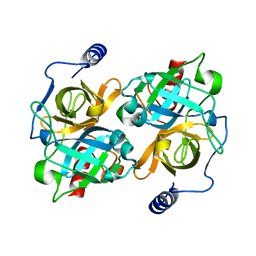 | | New insights into the P186 flip and oligomeric state of Staphylococcus aureus exfoliative toxin E: implications for the exfoliative mechanism | | Descriptor: | Exfoliative toxin E | | Authors: | Gismene, C, Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, Santisteban, A.R.N, de Moraes, F.R, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Staphylococcus aureus Exfoliative Toxin E, Oligomeric State and Flip of P186: Implications for Its Action Mechanism.
Int J Mol Sci, 23, 2022
|
|
7ZZX
 
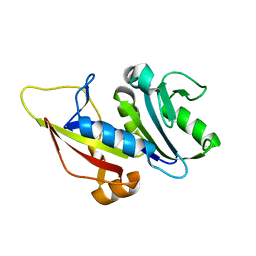 | |
6Y8B
 
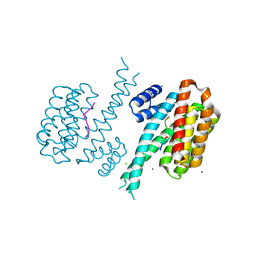 | | 14-3-3 Sigma in complex with phosphorylated caspase{pS139} peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7PJJ
 
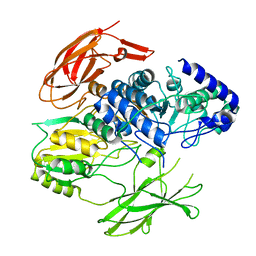 | | Structure of the Family-3 Glycosyl Hydrolase BcpE2 from Streptomyces scabies | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Jadot, C, Herman, R, Deflandre, B, Rigali, S, Kerff, F. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Structure and Function of BcpE2, the Most Promiscuous GH3-Family Glucose Scavenging Beta-Glucosidase.
Mbio, 13, 2022
|
|
6YBI
 
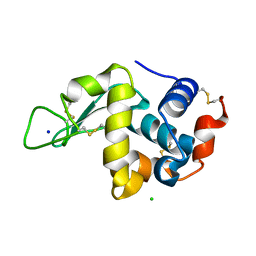 | |
6YBO
 
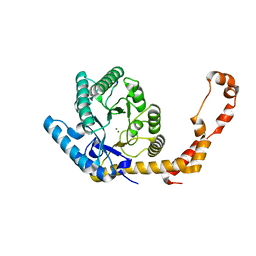 | |
6YBX
 
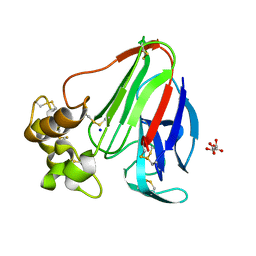 | |
6Y07
 
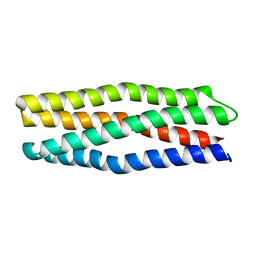 | |
6Y3M
 
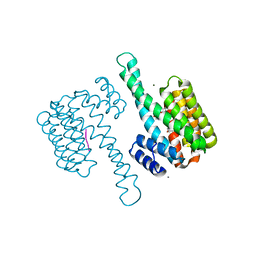 | | 14-3-3 Sigma in complex with phosphorylated ATPase peptide | | Descriptor: | 14-3-3 protein sigma, ATPase peptide, CALCIUM ION, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6Y8E
 
 | | 14-3-3 Sigma in complex with phosphorylated MLF1 peptide | | Descriptor: | 14-3-3 protein sigma, ARG-SER-PHE-SEP-GLU-PRO-PHE-GLY, CALCIUM ION, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6Y4K
 
 | |
7PX1
 
 | | Conotoxin from Conus mucronatus | | Descriptor: | CADMIUM ION, Conus mucronatus, IODIDE ION | | Authors: | Mueller, E, Hackney, C.M, Ellgaard, L, Morth, J.P. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A previously unrecognized superfamily of macro-conotoxins includes an inhibitor of the sensory neuron calcium channel Cav2.3.
Plos Biol., 21, 2023
|
|
6Y8Y
 
 | | Structure of Baltic Herring (Clupea Harengus) Phosphoglucomutase 5 (PGM5) with bound Glucose-1-Phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Gustafsson, R, Eckhard, U, Selmer, M. | | Deposit date: | 2020-03-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring-An Inactive Enzyme with Intact Substrate Binding.
Biomolecules, 10, 2020
|
|
