3I4C
 
 | | Crystal structure of Sulfolobus Solfataricus ADH(SsADH) double mutant (W95L,N249Y) | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Pennacchio, A, Zagari, A, Rossi, M, Raia, C.A. | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Tryptophan 95 in substrate specificity and structural stability of Sulfolobus solfataricus alcohol dehydrogenase
Extremophiles, 13, 2009
|
|
6IGY
 
 | |
1DX6
 
 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-GALANTHAMINE AT 2.3A RESOLUTION | | Descriptor: | (-)-GALANTHAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Greenblatt, H.M, Kryger, G, Lewis, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 1999-12-21 | | Release date: | 2000-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Acetylcholinesterase Complexed with (-)-Galanthamine at 2.3A Resolution
FEBS Lett., 463, 1999
|
|
2WX5
 
 | | Hexa-coordination of a bacteriochlorophyll cofactor in the Rhodobacter sphaeroides reaction centre | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Marsh, M, Frolov, D, Crouch, L.I, Fyfe, P.K, Robert, B, van Grondelle, R, Jones, M.R, Hadfield, A.T. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Spectroscopic Consequences of Hexa-Coordination of a Bacteriochlorophyll Cofactor in the Rhodobacter Sphaeroides Reaction Centre
Biochemistry, 49, 2010
|
|
1CMT
 
 | | THE ROLE OF ASPARTATE-235 IN THE BINDING OF CATIONS TO AN ARTIFICIAL CAVITY AT THE RADICAL SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Trester, M.L, Jensen, G.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1995-04-11 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of aspartate-235 in the binding of cations to an artificial cavity at the radical site of cytochrome c peroxidase.
Protein Sci., 4, 1995
|
|
2V0A
 
 | | Atomic resolution crystal structure of Human Superoxide Dismutase | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S, Yong, C.W, Smith, W, Hasnain, S.S. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Dynamics Using Atomic-Resolution Structure Reveal Structural Fluctuations that May Lead to Polymerization of Human Cu-Zn Superoxide Dismutase.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5DCE
 
 | |
2NNX
 
 | |
2O92
 
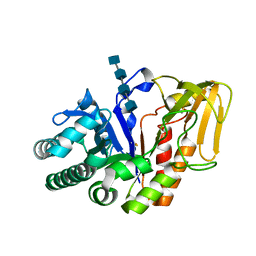 | | Crystal structure of a signalling protein (SPG-40) complex with tetrasaccharide at 3.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sharma, P, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-12-13 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a signalling protein (SPG-40) complex with tetrasaccharide at 3.0A resolution
To be Published
|
|
8A1E
 
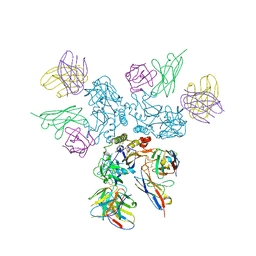 | | Rabies virus glycoprotein in complex with Fab fragments of 17C7 and 1112-1 neutralizing antibodies | | Descriptor: | Fab 1112-1 heavy chain variable domain, Fab 1112-1 light chain variable domain, Fab 17C7 heavy chain variable domain, ... | | Authors: | Ng, W.M, Fedosyuk, S, English, S, Augusto, G, Berg, A, Thorley, L, Haselon, A.S, Segireddy, R.R, Bowden, T.A, Douglas, A.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of trimeric pre-fusion rabies virus glycoprotein in complex with two protective antibodies.
Cell Host Microbe, 30, 2022
|
|
7ZQ9
 
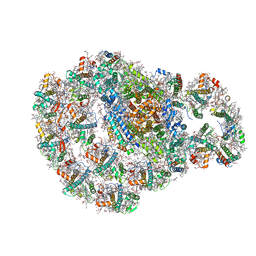 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
1JPS
 
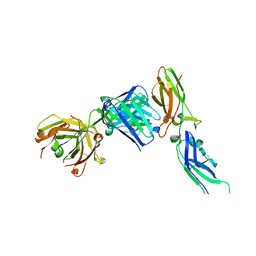 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
7ZKP
 
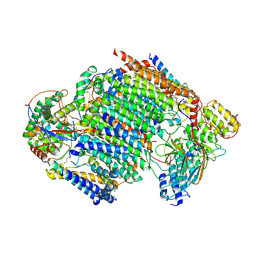 | | Late assembly intermediate of the proximal proton pumping module of complex I with assembly factors NDUFAF1 and CIA84 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, ... | | Authors: | Schiller, J, Laube, E, Vonck, J, Zickermann, V. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-23 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into complex I assembly: Function of NDUFAF1 and a link with cardiolipin remodeling.
Sci Adv, 8, 2022
|
|
1JTA
 
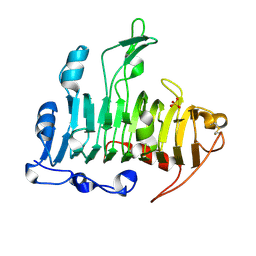 | | Crystal Structure of Pectate Lyase A (C2 form) | | Descriptor: | SULFATE ION, pectate lyase A | | Authors: | Thomas, L.M, Doan, C.N, Oliver, R.L, Yoder, M.D. | | Deposit date: | 2001-08-20 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of pectate lyase A: comparison to other isoforms.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7ZQC
 
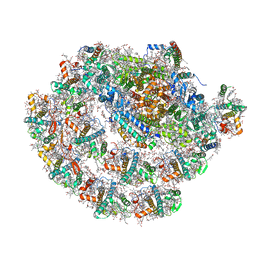 | | Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
1JMC
 
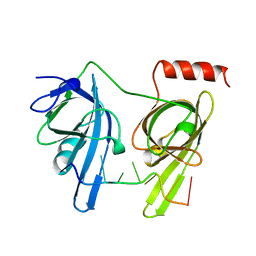 | | SINGLE STRANDED DNA-BINDING DOMAIN OF HUMAN REPLICATION PROTEIN A BOUND TO SINGLE STRANDED DNA, RPA70 SUBUNIT, RESIDUES 183-420 | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*C)-3'), PROTEIN (REPLICATION PROTEIN A (RPA)) | | Authors: | Bochkarev, A, Pfuetzner, R, Edwards, A, Frappier, L. | | Deposit date: | 1996-11-11 | | Release date: | 1997-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA.
Nature, 385, 1997
|
|
7ZQD
 
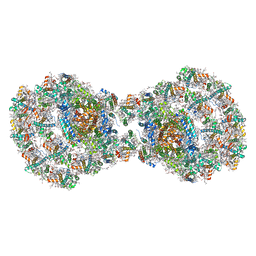 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZWD
 
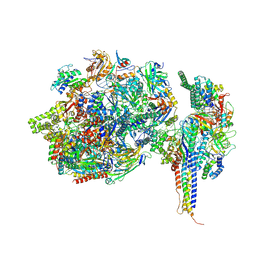 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1JN0
 
 | | Crystal structure of the non-regulatory A4 isoform of spinach chloroplast glyceraldehyde-3-phosphate dehydrogenase complexed with NADP | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Fermani, S, Ripamonti, A, Sabatino, P, Zanotti, G, Scagliarini, S, Sparla, F, Trost, P, Pupillo, P. | | Deposit date: | 2001-07-21 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the non-regulatory A(4 )isoform of spinach chloroplast glyceraldehyde-3-phosphate dehydrogenase complexed with NADP.
J.Mol.Biol., 314, 2001
|
|
7ZWC
 
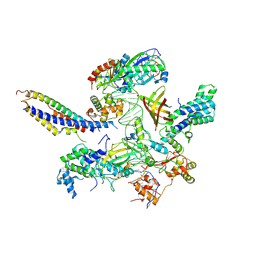 | | Structure of SNAPc:TBP-TFIIA-TFIIB sub-complex bound to U5 snRNA promoter | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX7
 
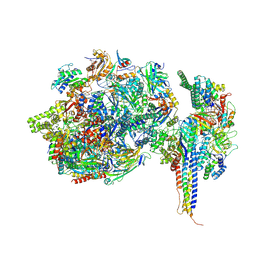 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8A5Q
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on straight DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5A
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of INO80 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
1JZ4
 
 | |
