8D5S
 
 | |
8D7S
 
 | |
8D8B
 
 | |
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
7RFP
 
 | | Mouse GITR (mGITR) with DTA-1 Fab fragment | | Descriptor: | DTA-1 (heavy chain), DTA-1 (light chain), Tumor necrosis factor receptor superfamily member 18,Enhanced green fluorescent protein | | Authors: | Meyerson, J.R, He, C. | | Deposit date: | 2021-07-14 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism.
Sci Adv, 8, 2022
|
|
9D1X
 
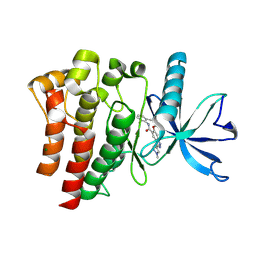 | | Crystal structure of FGFR3 bound to indazole inhibitor | | Descriptor: | (3P)-N-(2,6-dimethylphenyl)-6-methoxy-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazole-5-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3, ... | | Authors: | Postel, S, Johnson, Z.L. | | Deposit date: | 2024-08-08 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | OPLS5: Addition of Polarizability and Improved Treatment of Metals
Chemrxiv, 2024
|
|
8FXB
 
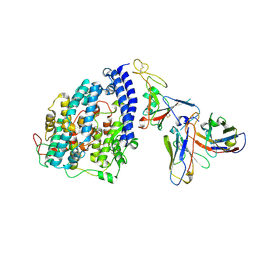 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
4RE5
 
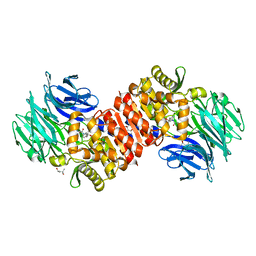 | | Acylaminoacyl peptidase complexed with a chloromethylketone inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Acylamino-acid-releasing enzyme, ... | | Authors: | Menyhard, D.K, Orgovan, Z, Szeltner, Z, Szamosi, I, Harmat, V. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytically distinct states captured in a crystal lattice: the substrate-bound and scavenger states of acylaminoacyl peptidase and their implications for functionality.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9DKC
 
 | | Structure of URAT1 in complex with TD-3 | | Descriptor: | 2-({1-[(4-bromonaphthalen-1-yl)methyl]-1H-imidazo[4,5-b]pyridin-2-yl}sulfanyl)-2-methylpropanoic acid, URAT1 | | Authors: | Suo, Y, Fedor, J.G, Lee, S.-Y. | | Deposit date: | 2024-09-08 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Molecular basis of the urate transporter URAT1 inhibition by gout drugs.
Nat Commun, 16, 2025
|
|
6WYA
 
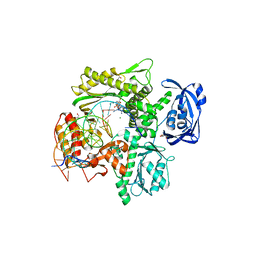 | | RTX (Reverse Transcription Xenopolymerase) in complex with a DNA duplex and dAMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FEC
 
 | | Structure of J-PKAc chimera complexed with Aplithianine derivative | | Descriptor: | 6-[(6P)-6-(4-bromo-1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7H-purine, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
6HKM
 
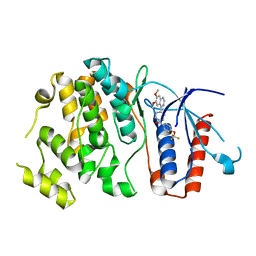 | | Crystal structure of Compound 1 with ERK5 | | Descriptor: | Mitogen-activated protein kinase 7, [4-(6,7-dimethoxyquinazolin-4-yl)piperidin-1-yl]-[4-(trifluoromethyloxy)phenyl]methanone | | Authors: | Nguyen, D, Lemos, C, Wortmann, L, Eis, K, Holton, S.J, Boemer, U, Lechner, C, Prechtl, S, Suelze, D, Siegel, F, Prinz, F, Lesche, R, Nicke, B, Mumberg, D, Bauser, M, Haegebarth, A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective (Piperidin-4-yl)pyrido[3,2- d]pyrimidine Based in Vitro Probe BAY-885 for the Kinase ERK5.
J. Med. Chem., 62, 2019
|
|
6X3J
 
 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F0224 (46) | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
4RI9
 
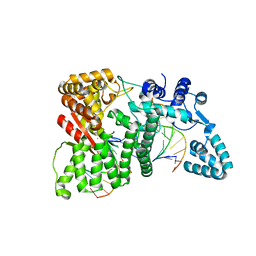 | |
4RIB
 
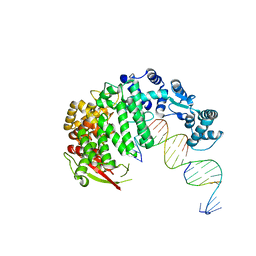 | | FAN1 Nuclease bound to 5' phosphorylated p(dT) single flap DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*GP*GP*AP*GP*TP*CP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*TP*GP*AP*GP*GP*CP*GP*TP*G)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
5GM7
 
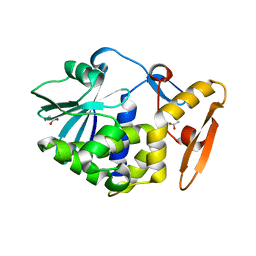 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina at 1.78 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Ribosome inactivating protein | | Authors: | Singh, B, Singh, P.K, Pandey, S.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-07-13 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina at 1.78 Angstrom resolution
To Be Published
|
|
8S1Y
 
 | | ThaOS V79A | | Descriptor: | (2~{R},4~{R})-5,5-dimethyl-2-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-1,3-thiazolidine-4-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Basle, A, Ashley, B, Campopiano, D.J, Marles-wright, J. | | Deposit date: | 2024-02-16 | | Release date: | 2025-03-05 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rational engineering of a thermostable alpha-oxoamine synthase biocatalyst expands the substrate scope and synthetic applicability.
Commun Chem, 8, 2025
|
|
8YW7
 
 | | Structure of CylI in complex with resorcinol product | | Descriptor: | 5-[(2~{S},7~{R})-7-chloranyl-2-methyl-octyl]benzene-1,3-diol, CylI | | Authors: | Wang, H.Q, Xiang, Z. | | Deposit date: | 2024-03-30 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into type III polyketide synthase CylI from cylindrocyclophane biosynthesis.
Protein Sci., 33, 2024
|
|
7RON
 
 | |
8U1P
 
 | | Local refinement of Plasmodium falciparum gametocyte surface protein Pfs48/45 Domains 1 and 2 in complex with neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gametocyte surface protein P45/48, ... | | Authors: | Kucharska, I, Hailemariam, S, Ivanochko, D, Rubinstein, J, Julien, J.P. | | Deposit date: | 2023-09-01 | | Release date: | 2025-03-12 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural elucidation of full-length Pfs48/45 in complex with potent monoclonal antibodies isolated from a naturally exposed individual.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4RI8
 
 | |
8DD5
 
 | | Crystal structure of KAT6A in complex with inhibitor CTx-648 (PF-9363) | | Descriptor: | 2,6-dimethoxy-N-{4-methoxy-6-[(1H-pyrazol-1-yl)methyl]-1,2-benzoxazol-3-yl}benzene-1-sulfonamide, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Greasley, S.E, Johnson, E, Brodsky, O. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Targeting KAT6A/KAT6B dependencies in breast cancer with a novel selective, orally bioavailable KAT6 inhibitor, CTx-648/PF-9363
To Be Published
|
|
4RJ3
 
 | | CDK2 with EGFR inhibitor compound 8 | | Descriptor: | 1-cyclopentyl-N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-1H-pyrrolo[3,2-c]pyridin-6-amine, ACETATE ION, Cyclin-dependent kinase 2 | | Authors: | Eigenbrot, C, Yin, J. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
4RIT
 
 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-07 | | Release date: | 2014-10-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
8S6I
 
 | | Crystal structure of the human CDKL2 kinase domain with Compound 9 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclin-dependent kinase-like 2, ... | | Authors: | Chen, X, Ni, X, Brooke, L, Bullock, A.N. | | Deposit date: | 2024-02-27 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Characterization of a Chemical Probe for Cyclin-Dependent Kinase-Like 2.
Acs Med.Chem.Lett., 15, 2024
|
|
