6RDM
 
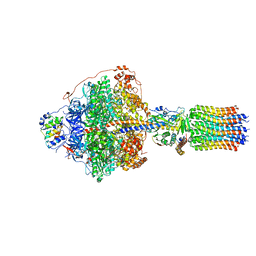 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1B, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDV
 
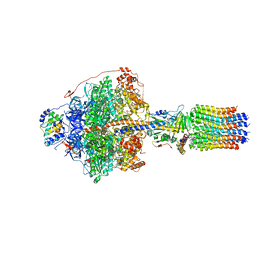 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
7BTT
 
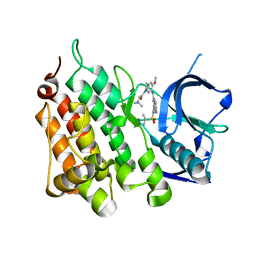 | | A X-ray cocrystal structure of XMU-MP-5 bound to the ALK kinase domain | | Descriptor: | 2-(dimethylamino)-1-[5-methoxy-6-[[4-[(2-propan-2-ylsulfonylphenyl)amino]-5H-pyrrolo[3,2-d]pyrimidin-2-yl]amino]-2,3-dihydroindol-1-yl]ethanone, ALK tyrosine kinase receptor | | Authors: | Yun, C.H, Zhu, S.J. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A X-ray cocrystal structure of XMU-MP-5 bound to the ALK kinase domain
To Be Published
|
|
9CGV
 
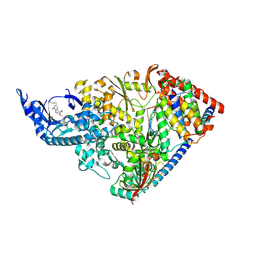 | | SARS-CoV-2 nsp12 NiRAN domain bound to a covalent inhibitor SW090466-1 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Hernandez, G, Tagliabracci, V.S. | | Deposit date: | 2024-07-01 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Covalent inhibition of the SARS-CoV-2 NiRAN domain via an active-site cysteine.
J.Biol.Chem., 301, 2025
|
|
5IXC
 
 | | Human GIVD cytosolic phospholipase A2 in complex with Methyl gamma-Linolenyl Fluorophosphonate | | Descriptor: | BARIUM ION, Cytosolic phospholipase A2 delta, methyl (R)-(6Z,9Z,12Z)-octadeca-6,9,12-trien-1-ylphosphonofluoridate | | Authors: | Wang, H, Klein, M.G. | | Deposit date: | 2016-03-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Human GIVD Cytosolic Phospholipase A2 Reveals Insights into Substrate Recognition.
J.Mol.Biol., 428, 2016
|
|
8SLS
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and ambient pressure (0.1 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8P5H
 
 | | Kinase domain of mutant human ULK1 in complex with compound CCT241533 | | Descriptor: | 4-FLUORO-2-(4-{[(3S,4R)-4-(1-HYDROXY-1-METHYLETHYL)PYRROLIDIN-3-YL]AMINO}-6,7-DIMETHOXYQUINAZOLIN-2-YL)PHENOL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
8BFJ
 
 | | CNOT11-GGNBP2 complex | | Descriptor: | 1,2-ETHANEDIOL, CCR4-NOT transcription complex subunit 11, Gametogenetin-binding protein 2 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8EXM
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with a JAK3 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK3 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8P5G
 
 | | Kinase domain of wild type human ULK1 in complex with compound CCT241533 | | Descriptor: | 4-FLUORO-2-(4-{[(3S,4R)-4-(1-HYDROXY-1-METHYLETHYL)PYRROLIDIN-3-YL]AMINO}-6,7-DIMETHOXYQUINAZOLIN-2-YL)PHENOL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
9CCI
 
 | |
6I7D
 
 | | Plasmodium falciparum Myosin A, post-rigor and rigor-like states | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Myosin-A | | Authors: | Robert-Paganin, J, Auguin, D, Moussaoui, D, Jousset, G, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism.
Nat Commun, 10, 2019
|
|
6X9O
 
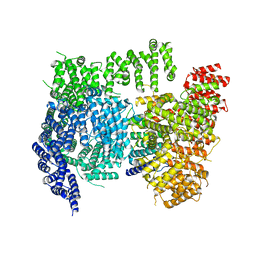 | | High resolution cryoEM structure of huntingtin in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Lea, S.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Commun Biol, 4, 2021
|
|
8P5K
 
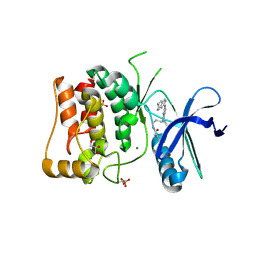 | | Kinase domain of mutant human ULK1 in complex with compound MRT68921 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
6R45
 
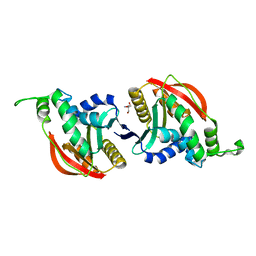 | | Crystal structure of eukaryotic O-GlcNAcase HAT-like domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, O-GlcNAcase | | Authors: | Raimi, O.G, Gorelik, A, Hopkins-Navratilova, I, Aristotelous, T, Ferenbach, A, vanAalten, D. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Crystal structure of eukaryotic O-GlcNAcase HAT-like domain
To Be Published
|
|
8SW0
 
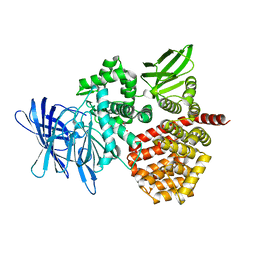 | | Puromycin sensitive aminopeptidase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Puromycin-sensitive aminopeptidase, ... | | Authors: | Rodgers, D.W, Sampath, S. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of puromycin-sensitive aminopeptidase and polyglutamine binding.
Plos One, 18, 2023
|
|
8EYB
 
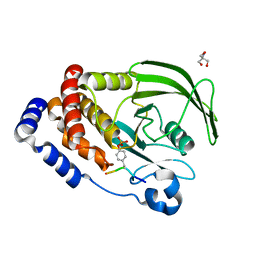 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2 activation loop phosphopeptide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8B70
 
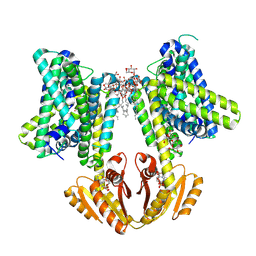 | | KimA from B. subtilis with nucleotide second-messenger c-di-AMP bound | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, DODECYL-BETA-D-MALTOSIDE, POTASSIUM ION, ... | | Authors: | Vonck, J, Wieferig, J.P. | | Deposit date: | 2022-09-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cyclic di-AMP traps proton-coupled K + transporters of the KUP family in an inward-occluded conformation.
Nat Commun, 14, 2023
|
|
8H0I
 
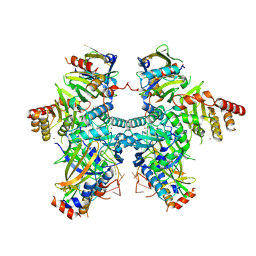 | | Cryo-EM structure of APOBEC3G-Vif complex | | Descriptor: | APOBEC3G, CHLORIDE ION, Core binding factor beta, ... | | Authors: | Kouno, T, Shibata, S, Hyun, J, Kim, T.G, Wolf, M. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into RNA bridging between HIV-1 Vif and antiviral factor APOBEC3G.
Nat Commun, 14, 2023
|
|
8P3C
 
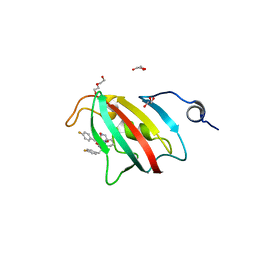 | | Full length structure of BpMIP with bound inhibitor NJS227. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-(pyridin-3-ylmethylamino)propan-2-yl]piperidine-2-carboxamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J.Med.Chem., 68, 2025
|
|
7SX5
 
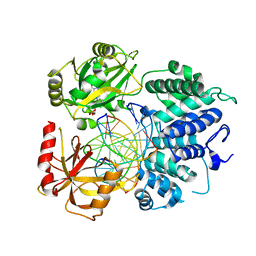 | | Crystal structure of ligase I with nick duplexes containing mismatch A:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA chain 1, DNA chain 2, ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
7D82
 
 | |
6RE1
 
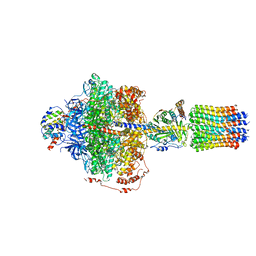 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2A, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
5N84
 
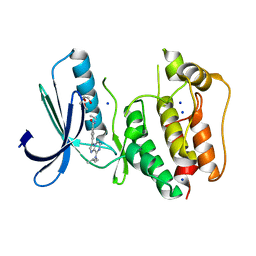 | | TTK kinase domain in complex with Mps-BAY2b | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Dual specificity protein kinase TTK, SODIUM ION, ... | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
8R12
 
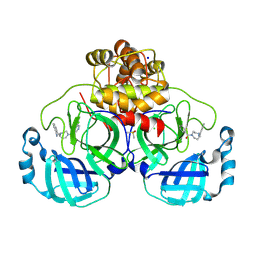 | | Structure of compound 8 bound to SARS-CoV-2 main protease | | Descriptor: | 2-[[4-(5-chloranylpyridin-3-yl)carbonyl-1,4-diazepan-1-yl]methyl]benzenecarbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
