3N7B
 
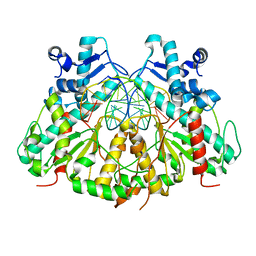 | | SgrAI bound to secondary site DNA and Ca(II) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*GP*TP*CP*CP*AP*CP*CP*GP*GP*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*CP*CP*CP*CP*CP*GP*GP*TP*GP*GP*AP*CP*T)-3'), ... | | Authors: | Horton, N.C, Little, E.J, Dunten, P.W. | | Deposit date: | 2010-05-26 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | New clues in the allosteric activation of DNA cleavage by SgrAI: structures of SgrAI bound to cleaved primary-site DNA and uncleaved secondary-site DNA.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4IJ0
 
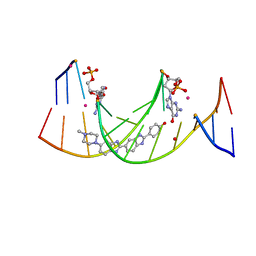 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
1LEB
 
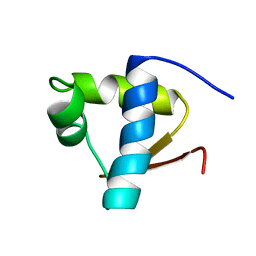 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
3KNT
 
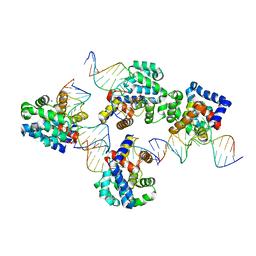 | |
4OD6
 
 | | Structure of Smr domain of MutS2 from Deinococcus radiodurans, Mn2+ soaked | | Descriptor: | Endonuclease MutS2 | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
8REV
 
 | |
6FAS
 
 | | Crystal structure of VAL1 B3 domain in complex with cognate DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*GP*CP*CP*AP*TP*GP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*GP*CP*AP*TP*GP*GP*CP*T)-3') | | Authors: | Sasnauskas, G. | | Deposit date: | 2017-12-17 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1.
Nucleic Acids Res., 46, 2018
|
|
1MGT
 
 | | CRYSTAL STRUCTURE OF O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM HYPERTHERMOPHILIC ARCHAEON PYROCOCCUS KODAKARAENSIS STRAIN KOD1 | | Descriptor: | PROTEIN (O6-METHYLGUANINE-DNA METHYLTRANSFERASE), SULFATE ION | | Authors: | Hashimoto, H, Inoue, T, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase.
J.Mol.Biol., 292, 1999
|
|
5CDN
 
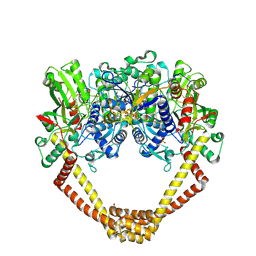 | | 2.8A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP**GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
7WCG
 
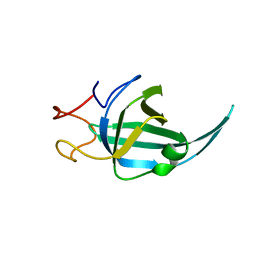 | |
1MTL
 
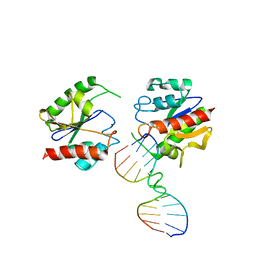 | | Non-productive MUG-DNA complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Savva, R, Barlow, T, Brown, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 2002-09-21 | | Release date: | 2002-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct.
Nat.Struct.Biol., 5, 1998
|
|
6FB0
 
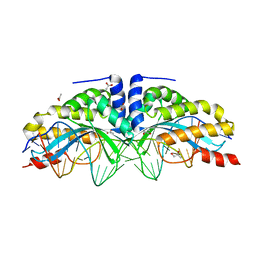 | |
5CDQ
 
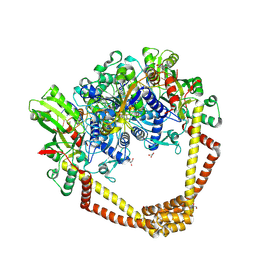 | | 2.95A structure of Moxifloxacin with S.aureus DNA gyrase and DNA | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CG9
 
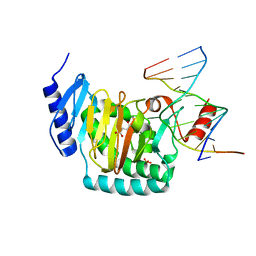 | | NgTET1 in complex with 5mC DNA in space group P3221 | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DNA (5'-D(*TP*GP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
5CG8
 
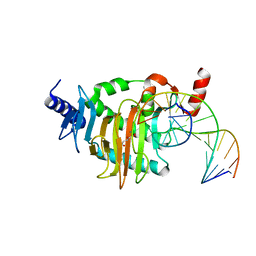 | | NgTET1 in complex with 5hmC DNA | | Descriptor: | 2-OXOGLUTARIC ACID, DNA (5'-D(*AP*GP*AP*AP*TP*TP*CP*CP*GP*TP*TP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*(5HC)P*GP*GP*AP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
5CDO
 
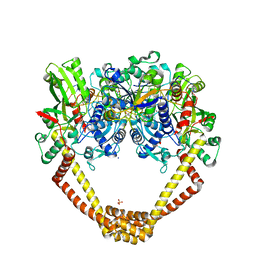 | | 3.15A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-4',6'-dihydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidin]-2'-one, (2R,4S,4aS,5R)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, (2R,4S,4aS,5S)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
6GDS
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | Telomeric DNA (5' CTAACCCTAA) 10mer, Telomeric DNA (5'-TTAGGGTTAG)-3') 10mer | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
1LEA
 
 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
5VVK
 
 | | Cas1-Cas2 bound to full-site mimic | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*CP*CP*AP*CP*CP*AP*GP*TP*G)-3'), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.D, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
2MRU
 
 | | Structure of truncated EcMazE-DNA complex | | Descriptor: | Antitoxin MazE, DNA (5'-D(*CP*GP*TP*GP*AP*TP*AP*TP*AP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*CP*TP*AP*TP*AP*TP*AP*TP*CP*AP*CP*G)-3') | | Authors: | Zorzini, V, Buts, L, Loris, R, van Nuland, N. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Escherichia coli antitoxin MazE as transcription factor: insights into MazE-DNA binding.
Nucleic Acids Res., 43, 2015
|
|
5U30
 
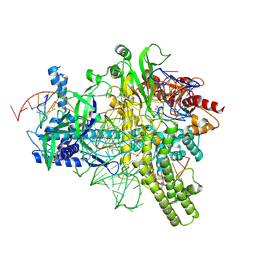 | | Crystal structure of AacC2c1-sgRNA-extended target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
1D32
 
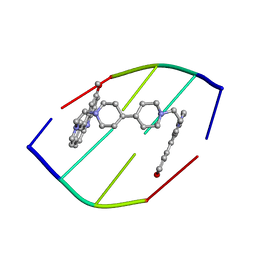 | | DRUG-INDUCED DNA REPAIR: X-RAY STRUCTURE OF A DNA-DITERCALINIUM COMPLEX | | Descriptor: | DITERCALINIUM, DNA (5'-D(*CP*GP*CP*G)-3') | | Authors: | Gao, Q, Williams, L.D, Egli, M, Rabinovich, D, Chen, S.-L, Quigley, G.J, Rich, A. | | Deposit date: | 1991-01-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Drug-induced DNA repair: X-ray structure of a DNA-ditercalinium complex.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
5U31
 
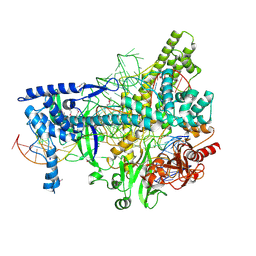 | | Crystal structure of AacC2c1-sgRNA-8mer substrate DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
4OCH
 
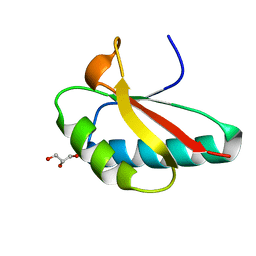 | | Apo structure of Smr domain of MutS2 from Deinococcus radiodurans | | Descriptor: | Endonuclease MutS2, GLYCEROL | | Authors: | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4001 Å) | | Cite: | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
3ODH
 
 | | Structure of OkrAI/DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*TP*GP*GP*AP*TP*CP*CP*AP*TP*A)-3'), OkrAI endonuclease | | Authors: | Vanamee, E.S, Aggarwal, A.K. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asymmetric DNA recognition by the OkrAI endonuclease, an isoschizomer of BamHI.
Nucleic Acids Res., 39, 2011
|
|
