8SMX
 
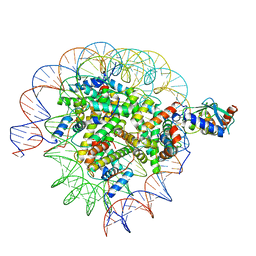 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SMW
 
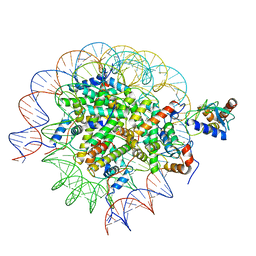 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SMV
 
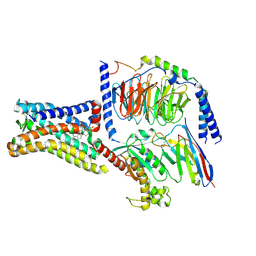 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Biorxiv, 2023
|
|
8SMU
 
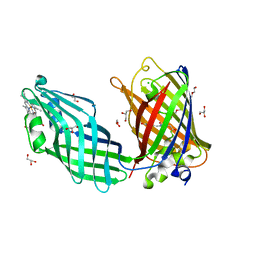 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
8SMT
 
 | | Crystal structure of antibody WRAIR-2134 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2134 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SMS
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase, FabB, and cerulenin crosslinker-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-{2-[(2R)-2-hydroxy-4-oxododecanamido]ethyl}-beta-alaninamide | | Authors: | Jiang, Z, Chen, A, Chen, J, Sekhon, A, Louie, G.V, Noel, J.P, La Clair, J.J, Burkart, M.D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Masked cerulenin enables a dual-site selective protein crosslink.
Chem Sci, 14, 2023
|
|
8SMR
 
 | | cytochrome bc1-cbb3 supercomplex from Pseudomonas aeruginosa | | Descriptor: | (2R)-2-(methoxymethyl)-4-{[(25R)-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Di Trani, J.M, Rubinstein, J.L. | | Deposit date: | 2023-04-26 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the bc 1 - cbb 3 respiratory supercomplex from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
8SMP
 
 | | Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state | | Descriptor: | Fab15 heavy chain, Fab15 light chain, Hyaluronan synthase 1, ... | | Authors: | Gorniak, I, Zimmer, J. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomistic insights into hyaluronan synthesis, secretion, and length control
To Be Published
|
|
8SMO
 
 | |
8SMN
 
 | | Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab15 heavy chain, Fab15 light chain, ... | | Authors: | Gorniak, I, Zimmer, J. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomistic insights into hyaluronan synthesis, secretion, and length control
To Be Published
|
|
8SMM
 
 | | Xenopus laevis hyaluronan synthase 1 | | Descriptor: | Fab15 heavy chain, Fab15 light chain, Hyaluronan synthase 1 | | Authors: | Gorniak, I, Zimmer, J. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomistic insights into hyaluronan synthesis, secretion, and length control
To Be Published
|
|
8SML
 
 | | hPAD4 bound to inhibitory Fab hI365 | | Descriptor: | CALCIUM ION, Fab hI365 heavy chain, Fab hI365 light chain, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8SMK
 
 | | hPAD4 bound to Activating Fab hA362 | | Descriptor: | Activating Fab 362 heavy chain, Activating Fab 362 light chain, CALCIUM ION, ... | | Authors: | Maker, A, Verba, K.A. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8SMJ
 
 | |
8SMI
 
 | | Crystal structure of antibody WRAIR-2123 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2123 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SMH
 
 | |
8SMG
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMF
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Gp34.65, MAGNESIUM ION | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SME
 
 | | Structure of SPO1 phage Tad2 in apo state | | Descriptor: | Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMD
 
 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, ABC transporter ATPase | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
8SMC
 
 | | Cryo-EM structure of LRRK2 bound with type-I inhibitor DNL201 | | Descriptor: | 2-methyl-2-(3-methyl-4-{[4-(methylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1H-pyrazol-1-yl)propanenitrile, GUANOSINE-5'-DIPHOSPHATE, non-specific serine/threonine protein kinase | | Authors: | Sun, J, Zhu, H. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Rab29-dependent asymmetrical activation of leucine-rich repeat kinase 2.
Science, 382, 2023
|
|
8SMA
 
 | | SfbO with di-Manganese cofactor | | Descriptor: | Amidohydrolase family protein, MANGANESE (II) ION, SULFATE ION | | Authors: | Liu, C, Rittle, J. | | Deposit date: | 2023-04-25 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bioinformatic Discovery of a Cambialistic Monooxygenase.
J.Am.Chem.Soc., 146, 2024
|
|
8SM9
 
 | |
8SM8
 
 | |
