7RYU
 
 | |
1FAX
 
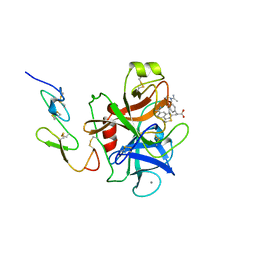 | | COAGULATION FACTOR XA INHIBITOR COMPLEX | | Descriptor: | (2S)-3-(7-carbamimidoylnaphthalen-2-yl)-2-[4-({(3R)-1-[(1Z)-ethanimidoyl]pyrrolidin-3-yl}oxy)phenyl]propanoic acid, CALCIUM ION, FACTOR XA | | Authors: | Brandstetter, H, Engh, R.A. | | Deposit date: | 1996-08-23 | | Release date: | 1997-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of active site-inhibited clotting factor Xa. Implications for drug design and substrate recognition.
J.Biol.Chem., 271, 1996
|
|
8FZY
 
 | |
1FIR
 
 | |
8B16
 
 | |
8WWQ
 
 | | geniposidic acid O-methyltransferase complexed with SAH and geniposidic acid | | Descriptor: | 1,2-ETHANEDIOL, Geniposidic acid, MAGNESIUM ION, ... | | Authors: | Luo, Z, Li, L, Ma, D, Zhou, H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the substrate specificity of GAMT and divergence of seco-iridoid and closed-ring iridoid
To Be Published
|
|
303D
 
 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL OUT' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | Descriptor: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | Deposit date: | 1996-06-26 | | Release date: | 1997-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
1G8F
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1FU0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE PHOSPHO-SERINE 46 HPR FROM ENTEROCOCCUS FAECALIS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Audette, G.F, Engelmann, R, Hengstenberg, W, Deutscher, J, Hayakawa, K, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A resolution structure of phospho-serine 46 HPr from Enterococcus faecalis.
J.Mol.Biol., 303, 2000
|
|
1GHV
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
7MNK
 
 | | Crystal structure of the tetramerization element of NUP358/RanBP2 (residues 805-832) | | Descriptor: | 1,2-ETHANEDIOL, E3 SUMO-protein ligase RanBP2, SULFATE ION | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7RHA
 
 | | A new fluorescent protein darkmRuby at pH 5.0 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new fluorescent protein darkmRuby at pH 5.0
To Be Published
|
|
6R9K
 
 | |
8UX2
 
 | | Chromobacterium violaceum mono-ADP-ribosyltransferase CteC in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NAD(+)--protein-threonine ADP-ribosyltransferase, ... | | Authors: | Zhang, Z, Rondon, H, Das, C. | | Deposit date: | 2023-11-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of bacterial ubiquitin ADP-ribosyltransferase CteC reveals a substrate-recruiting insertion.
J.Biol.Chem., 300, 2023
|
|
6Z2D
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 41 21 2 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, O-glycan protease, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-15 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
7S6G
 
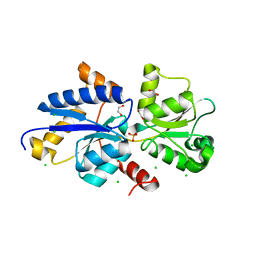 | | Crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Marine picocyanobacterial PhnD1 shows specificity for various phosphorus sources but likely represents a constitutive inorganic phosphate transporter.
Isme J, 17, 2023
|
|
8FXP
 
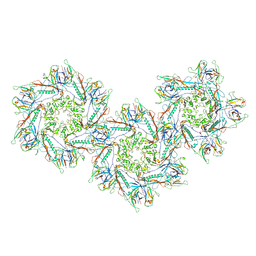 | | Structure of capsid of Agrobacterium phage Milano | | Descriptor: | Linking protein 1, gp16, Linking protein 2, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-25 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
7SFJ
 
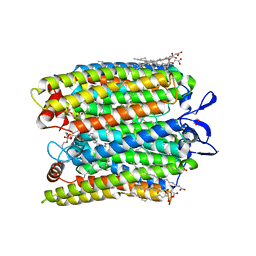 | | ChRmine in MSP1E3D1 lipid nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ChRmine, RETINAL | | Authors: | Tucker, K, Brohawn, S. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of the channelrhodopsin ChRmine in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7SFK
 
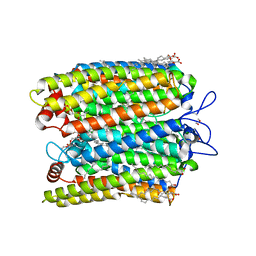 | | ChRmine in MSP1E3D1 lipid nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ChRmine, RETINAL | | Authors: | Tucker, K, Brohawn, S. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of the channelrhodopsin ChRmine in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
6R9L
 
 | |
6FYS
 
 | | Structure of single domain antibody SD83 | | Descriptor: | 1,2-ETHANEDIOL, Single domain antibody SD83 | | Authors: | Laursen, N.S, Wilson, I.A. | | Deposit date: | 2018-03-12 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
1V9G
 
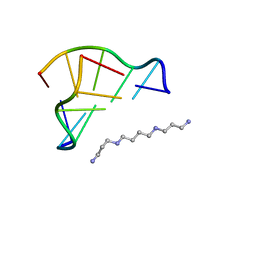 | | Neutron Crystallographic analysis of the Z-DNA hexamer CGCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', N,N'-BIS(3-AMMONIOPROPYL)BUTANE-1,4-DIAMINIUM | | Authors: | Chatake, T, Tanaka, I, Niimura, N. | | Deposit date: | 2004-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | The hydration structure of a Z-DNA hexameric duplex determined by a neutron diffraction technique.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5JWZ
 
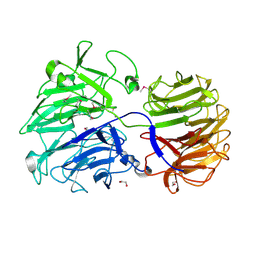 | |
302D
 
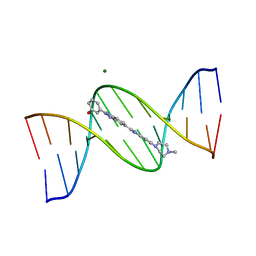 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL IN' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | Descriptor: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | Deposit date: | 1996-06-26 | | Release date: | 1997-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
9OG8
 
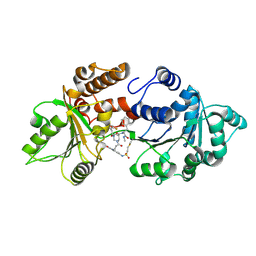 | | Crystal structure of WRN in complex with compound 43 | | Descriptor: | (3R,4S)-N-[(2R)-3-cyclohexyl-1-(methylamino)-1-oxopropan-2-yl]-4-{3-[6-(ethanesulfonamido)-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl]propanamido}-1-[(2-fluoro-4-methylphenyl)acetyl]piperidine-3-carboxamide, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION | | Authors: | Pemberton, O.A, Tong, Y, Nocek, B, Tang, H.Y.H. | | Deposit date: | 2025-04-30 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Validation, Key Pharmacophores, and X-ray Cocrystal Structures of Novel Biochemically and Cellularly Active WRN Inhibitors Derived from a DNA-Encoded Library Screen.
J.Med.Chem., 68, 2025
|
|
