6SCZ
 
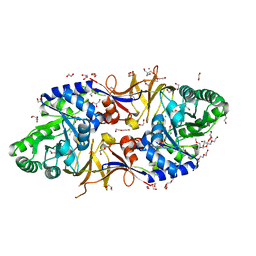 | | Mycobacterium tuberculosis alanine racemase inhibited by DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | de Chiara, C, Purkiss, A, Prosser, G, Homsak, M, de Carvalho, L.P.S. | | Deposit date: | 2019-07-26 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | D-Cycloserine destruction by alanine racemase and the limit of irreversible inhibition.
Nat.Chem.Biol., 16, 2020
|
|
4N49
 
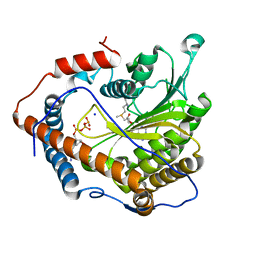 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with m7GpppG and SAM | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
7R7W
 
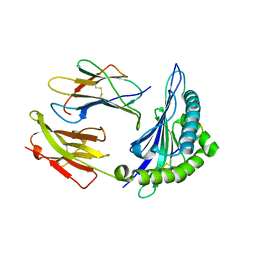 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, MHC class I antigen | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
4N6D
 
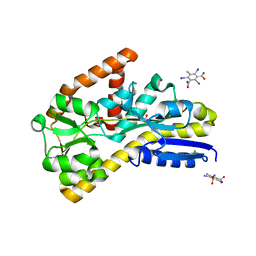 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638 (Desal_3247), Target EFI-510112, phased with I3C, open complex, C-terminus of symmetry mate bound in ligand binding site | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5EZZ
 
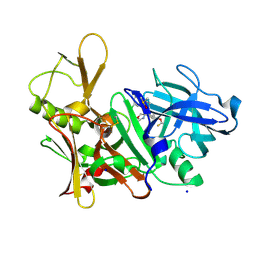 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH (4S)-4-[3-(5-chloro-3-pyridyl)phenyl]-4-[4-(difluoromethoxy)-3-methyl-phenyl]-5H-oxazol-2-amine | | Descriptor: | (4~{S})-4-[4-[bis(fluoranyl)methoxy]-3-methyl-phenyl]-4-[3-(5-chloranylpyridin-3-yl)phenyl]-5~{H}-1,3-oxazol-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
3MK2
 
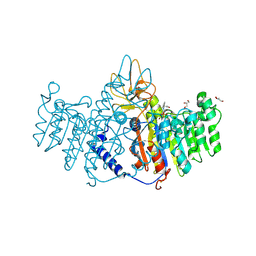 | | Placental alkaline phosphatase complexed with Phe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alkaline phosphatase, ... | | Authors: | Stec, B, Cheltsov, A, Millan, J.L. | | Deposit date: | 2010-04-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7R7X
 
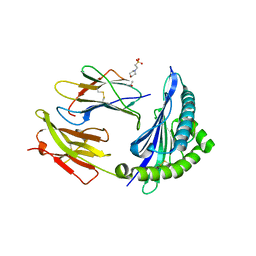 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7RWQ
 
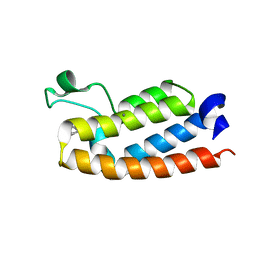 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
5F2F
 
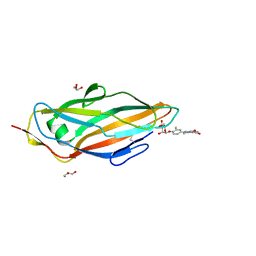 | | Crystal structure of para-biphenyl-2-methyl-3', 5' di-methyl amide mannoside bound to FimH lectin domain | | Descriptor: | 5-[4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-methyl-phenyl]-~{N}1,~{N}3-dimethyl-benzene-1,3-dicarboxamide, GLYCEROL, Protein FimH | | Authors: | Kalas, V, Hultgren, S.J. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Antivirulence Isoquinolone Mannosides: Optimization of the Biaryl Aglycone for FimH Lectin Binding Affinity and Efficacy in the Treatment of Chronic UTI.
Chemmedchem, 11, 2016
|
|
8PUN
 
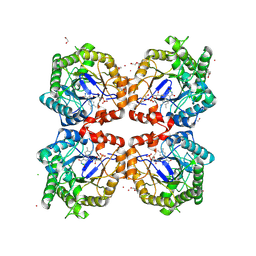 | | Old Yellow Enzyme from the thermophilic Ferrovum sp. JA12 | | Descriptor: | 1,2-ETHANEDIOL, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, CHLORIDE ION, ... | | Authors: | Polidori, N, Gruber, K. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Oligomerization, and Thermal Stability of a Recently Discovered Old Yellow Enzyme.
Proteins, 93, 2025
|
|
6D1Z
 
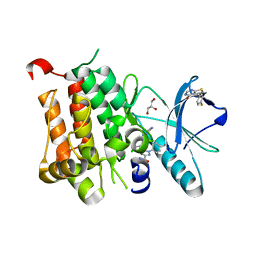 | | Crystal structure of Tyrosine-protein kinase receptor in complex with 5-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4(3H)-one Inhibitor | | Descriptor: | 5-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4(3H)-one, 5-{[5-(6-aminopyridin-2-yl)-2-chlorobenzene-1-carbonyl]amino}-1-phenyl-1H-pyrazole-3-carboxamide, GLYCEROL, ... | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Allosteric, Potent, Subtype Selective, and Peripherally Restricted TrkA Kinase Inhibitors.
J. Med. Chem., 62, 2019
|
|
1ZIV
 
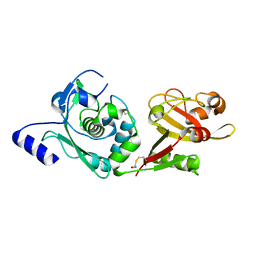 | | Catalytic Domain of Human Calpain-9 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain 9 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Dong, A, Choe, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition.
J.Mol.Biol., 366, 2007
|
|
4N9A
 
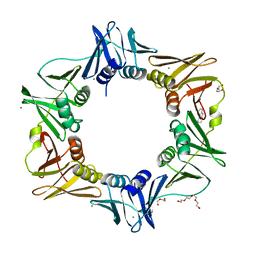 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
3MSD
 
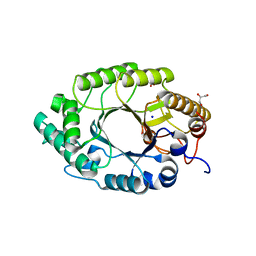 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
5F0H
 
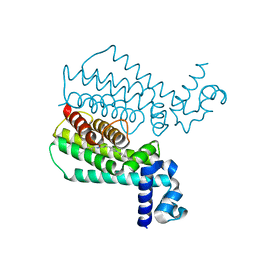 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 28 at 1.99A resolution | | Descriptor: | 3-[1-(4-cyanophenyl)piperidin-4-yl]-~{N}-[(4-piperidin-1-ylphenyl)methyl]propanamide, HTH-type transcriptional regulator EthR | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
8PH7
 
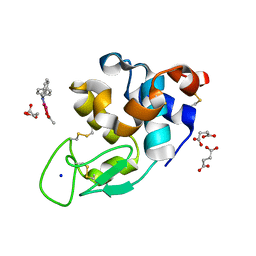 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DPhF)(O2CCH3)3] in condition A | | Descriptor: | Lysozyme C, SODIUM ION, SUCCINIC ACID, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
6DC6
 
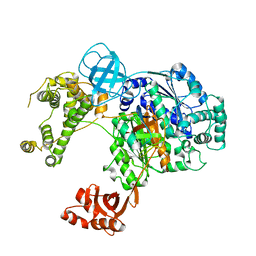 | | Crystal structure of human ubiquitin activating enzyme E1 (Uba1) in complex with ubiquitin | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, Ubiquitin, ... | | Authors: | Lv, Z, Yuan, L, Williams, K.M, Atkison, J.H, Olsen, S.K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of a human ubiquitin E1-ubiquitin complex reveals conserved functional elements essential for activity.
J. Biol. Chem., 293, 2018
|
|
5F36
 
 | |
6D55
 
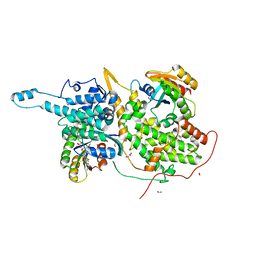 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-2-(2,6-diazaspiro[3.3]heptan-2-yl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-4-(4-methylpiperazin-1-yl)-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
2OYO
 
 | |
5EY3
 
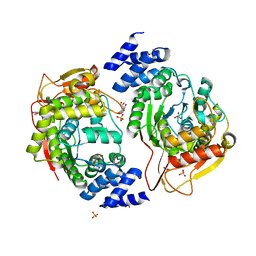 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with cytidine and sulphate | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with cytidine and sulphate
To Be Published
|
|
5EWX
 
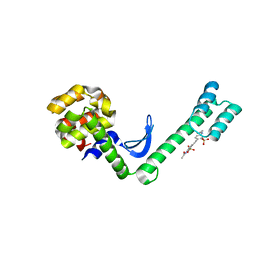 | | Fusion protein of T4 lysozyme and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Endolysin,Immunoglobulin G-binding protein A,Endolysin | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-11-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
4N5R
 
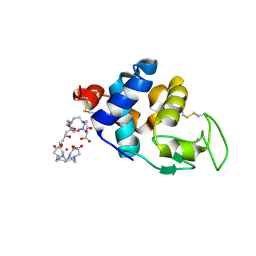 | | Hen egg-white lysozyme phased using free-electron laser data | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme C | | Authors: | Barends, T.R.M, Foucar, L, Botha, S, Doak, R.B, Shoeman, R.L, Nass, K, Koglin, J.E, Williams, G.J, Boutet, S, Messerschmidt, M, Schlichting, I. | | Deposit date: | 2013-10-10 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein crystal structure determination from X-ray free-electron laser data.
Nature, 505, 2014
|
|
7AD6
 
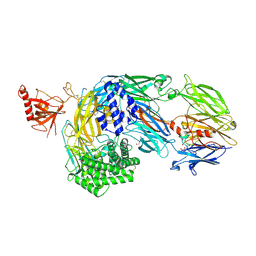 | | Crystal structure of human complement C5 in complex with the K92 bovine knob domain peptide. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYSTEINE, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
5F00
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-[3-[(3-chloro-8-quinolyl)amino]phenyl]-5-methyl-2,6-dihydro-1,4-oxazin-3-amine | | Descriptor: | (5~{R})-5-[3-[(3-chloranylquinolin-8-yl)amino]phenyl]-5-methyl-2,6-dihydro-1,4-oxazin-3-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
