2BVC
 
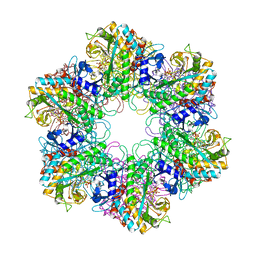 | | Crystal structure of Mycobacterium tuberculosis glutamine synthetase in complex with a transition state mimic | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLUTAMINE SYNTHETASE 1, ... | | Authors: | Krajewski, W.W, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2005-06-23 | | Release date: | 2005-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Mycobacterium Tuberculosis Glutamine Synthetase in Complex with a Transition-State Mimic Provides Functional Insights.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BXF
 
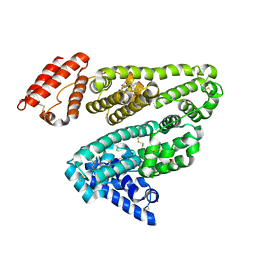 | | Human serum albumin complexed with diazepam | | Descriptor: | 7-CHLORO-1-METHYL-5-PHENYL-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
6VIU
 
 | |
5IN5
 
 | |
1SIS
 
 | | SPATIAL STRUCTURE OF INSECTOTOXIN I5A BUTHUS EUPEUS BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY (RUSSIAN) | | Descriptor: | SCORPION INSECTOTOXIN I5A | | Authors: | Arseniev, A.S, Kondakov, V.I, Lomize, A.L, Maiorov, V.N, Bystrov, V.F. | | Deposit date: | 1993-11-11 | | Release date: | 1994-04-30 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Determination of the spatial structure of insectotoxin 15A from Buthus erpeus by (1)H-NMR spectroscopy data
Bioorg.Khim., 17, 1991
|
|
5IP6
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-((tetrahydrofuran-3-yl)methyl)pyrrolidine-1-carboxamide at 1.93A resolution | | Descriptor: | N-{[(3S)-oxolan-3-yl]methyl}pyrrolidine-1-carboxamide, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
7MFV
 
 | | Crystal structure of synthetic nanobody (Sb16) | | Descriptor: | 1,2-ETHANEDIOL, Synthetic Nanobody #16 (Sb16) | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
5VW9
 
 | | Nicotinamide soak of Y316S mutant of corn root ferredoxin:NADP+ reductase in alternate space group | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
5VOH
 
 | | Crystal structure of engineered water-forming NADPH oxidase (TPNOX) bound to NADPH. The G159A, D177A, A178R, M179S, P184R mutant of LbNOX. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cracan, V, Grabarek, Z. | | Deposit date: | 2017-05-02 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | A genetically encoded tool for manipulation of NADP(+)/NADPH in living cells.
Nat. Chem. Biol., 13, 2017
|
|
5IRQ
 
 | |
5VJA
 
 | | Crystal Structure of human zipper-interacting protein kinase (ZIPK, alias DAPK3) in complex with a pyrazolo[3,4-d]pyrimidinone ligand (HS38) | | Descriptor: | (2R)-2-{[1-(3-chlorophenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]sulfanyl}propanamide, DIMETHYL SULFOXIDE, Death-associated protein kinase 3, ... | | Authors: | Carlson, D.A, Singer, M.R, Sutherland, C, Redondo, C, Alexander, L, Hughes, P.F, Knapp, S, MacDonald, J.A, Haystead, T.A.J. | | Deposit date: | 2017-04-19 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Targeting Pim Kinases and DAPK3 to Control Hypertension.
Cell Chem Biol, 25, 2018
|
|
7MSU
 
 | | Crystal structure of an archaeal CNNM, MtCorB, CBS-pair domain in complex with Mg2+-ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Hemolysin, ... | | Authors: | Chen, Y.S, Gehring, K. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of an archaeal CorB magnesium transporter.
Nat Commun, 12, 2021
|
|
5VN0
 
 | | Water-forming NADH oxidase from Lactobacillus brevis (LbNOX) bound to NADH. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cracan, V, Grabarek, Z. | | Deposit date: | 2017-04-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A genetically encoded tool for manipulation of NADP(+)/NADPH in living cells.
Nat. Chem. Biol., 13, 2017
|
|
3GSK
 
 | | A Bulky Rhodium Complex Bound to an Adenosine-Adenosine DNA Mismatch | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*AP*TP*TP*AP*CP*CP*G)-3', CACODYLATE ION, bis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)[chrysene-5,6-diiminato(2-)-kappa~2~N,N']rhodium(4+) | | Authors: | Zeglis, B.M, Pierre, V.C, Kaiser, J.T, Barton, J.K. | | Deposit date: | 2009-03-27 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode
Biochemistry, 48, 2009
|
|
4DYE
 
 | | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, isomerase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop
to be published
|
|
4DWF
 
 | |
3GYK
 
 | | The crystal structure of a thioredoxin-like oxidoreductase from Silicibacter pomeroyi DSS-3 | | Descriptor: | 1,2-ETHANEDIOL, 27kDa outer membrane protein, SULFATE ION | | Authors: | Fan, Y, Marshall, N, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-03 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The crystal structure of a thioredoxin-like oxidoreductase from Silicibacter pomeroyi DSS-3
To be Published
|
|
4LUP
 
 | | Crystal structure of the complex formed by region of E. coli sigmaE bound to its -10 element non template strand | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor, region 2 of sigmaE of E. coli | | Authors: | Campagne, S, Marsh, M.E, Vorholt, J.A.V, Allain, F.H.-T, Capitani, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for -10 promoter element melting by environmentally induced sigma factors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3TG7
 
 | | Crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution | | Descriptor: | Hexon protein | | Authors: | Zhu, Y, Roszak, A.W, Isaacs, N.W, McVey, J.H, Nicklin, S.A, Baker, A.H. | | Deposit date: | 2011-08-17 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution
To be Published
|
|
9C34
 
 | | Proline utilization A with the FADH- N5 atom covalently modified by proline | | Descriptor: | (2S)-3,4-dihydro-2H-pyrrole-2-carboxylic acid, (2~{S},5~{R})-5-[10-[(2~{S},3~{S},4~{R})-5-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-2,3,4-tris(oxidanyl)pentyl]-7,8-dimethyl-2,4-bis(oxidanylidene)benzo[g]pteridin-5-yl]pyrrolidine-2-carboxylic acid, Bifunctional protein PutA, ... | | Authors: | Tanner, J.J, Buckley, D.P. | | Deposit date: | 2024-05-31 | | Release date: | 2025-06-04 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Visualization of covalent intermediates and conformational states of proline utilization A by X-ray crystallography and molecular dynamics simulations.
J.Biol.Chem., 301, 2025
|
|
7M7E
 
 | | 6-Deoxyerythronolide B synthase (DEBS) hybrid module (M3/1) in complex with antibody fragment 1B2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
9C35
 
 | |
7M7H
 
 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1' | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
7M7F
 
 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
4LUV
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 1-(3-methylphenyl)-5-phenyl-1H-pyrazole-3-carboxylic acid, 5-(3-chloro-4-fluorophenyl)furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
