8T0Z
 
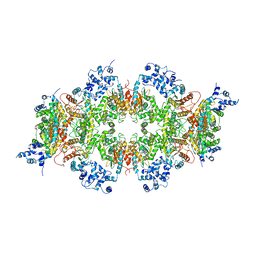 | | Human liver-type glutaminase (K253A) with L-Gln, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase liver isoform, mitochondrial | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-06-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
8T0Y
 
 | | TRPV1 in nanodisc bound with one LPA in one monomer | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, ... | | Authors: | Arnold, W.R, Cheng, Y. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8T0V
 
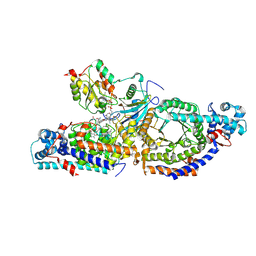 | | Closed state of lysine 5,6-aminomutase from Thermoanaerobacter tengcongensis | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-lysine 5,6-aminomutase alpha subunit, ... | | Authors: | Tian, S, Voss, P, Pham, K, Klose, T. | | Deposit date: | 2023-06-01 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Catalysis in Motion: Large-Scale Domain Shift Enables Co-C Bond Homolysis in Lysine 5,6-Aminomutase
To Be Published
|
|
8T0T
 
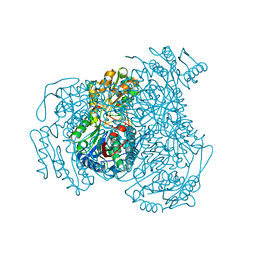 | | Structure of Compound 4 bound to human ALDH1A1 | | Descriptor: | 1-(4-{6-fluoro-3-[4-(methanesulfonyl)piperazine-1-carbonyl]quinolin-4-yl}phenyl)cyclopropane-1-carbonitrile, Aldehyde dehydrogenase 1A1, CHLORIDE ION, ... | | Authors: | Hurley, T.D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of substituted benzimidazoles as inhibitors of human aldehyde dehydrogenase 1A isoenzymes.
Chem.Biol.Interact., 391, 2024
|
|
8T0S
 
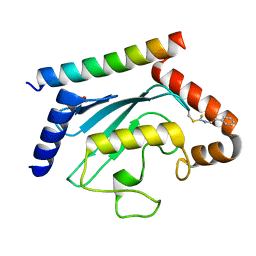 | | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Wang, C, Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position
To be published
|
|
8T0R
 
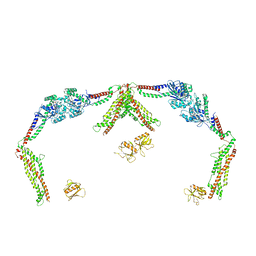 | |
8T0Q
 
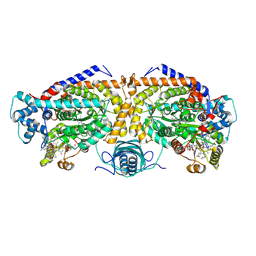 | | Open state of lysine 5,6-aminomutase from Thermoanaerobacter tengcongensis | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-lysine 5,6-aminomutase alpha subunit, ... | | Authors: | Tian, S, Voss, P, Pham, K, Klose, T. | | Deposit date: | 2023-06-01 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Catalysis in Motion: Large-Scale Domain Shift Enables Co-C Bond Homolysis in Lysine 5,6-Aminomutase
To Be Published
|
|
8T0P
 
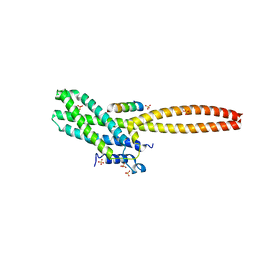 | | Structure of Cse4 bound to Ame1 and Okp1 | | Descriptor: | Histone H3-like centromeric protein CSE4, Inner kinetochore subunit AME1, Inner kinetochore subunit OKP1, ... | | Authors: | Deng, S, Harrison, S.C. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Recognition of centromere-specific histone Cse4 by the inner kinetochore Okp1-Ame1 complex.
Embo Rep., 24, 2023
|
|
8T0O
 
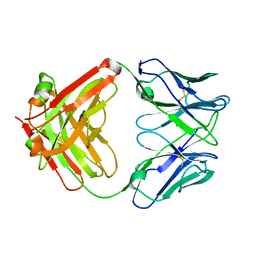 | | Fab from mAb RB2AT_87 | | Descriptor: | CHLORIDE ION, RB2AT_87 Fab Heavy chain, RB2AT_87 Fab Light chain | | Authors: | Kreutzer, A.G, Malonis, R.J, Lai, J.R, Nowick, J.S. | | Deposit date: | 2023-06-01 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Generation and Study of Antibodies against Two Triangular Trimers Derived from A beta.
Pept Sci (Hoboken), 116, 2024
|
|
8T0N
 
 | | Structure of Compound 4 bound to human ALDH1A1 | | Descriptor: | 2-methoxy-6-{[(1-propyl-1H-benzimidazol-2-yl)amino]methyl}phenol, Aldehyde dehydrogenase 1A1, CHLORIDE ION, ... | | Authors: | Hurley, T.D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of substituted benzimidazoles as inhibitors of human aldehyde dehydrogenase 1A isoenzymes.
Chem.Biol.Interact., 391, 2024
|
|
8T0M
 
 | | Proteasome 20S core particle from Pre1-1 Pre4-1 Double mutant | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Schnell, H, Velez, B, Hanna, J. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the preholoproteasome reveals late steps in proteasome core particle biogenesis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T0K
 
 | |
8T0J
 
 | | Salmonella Typhimurium ArnD | | Descriptor: | Probable 4-deoxy-4-formamido-L-arabinose-phosphoundecaprenol deformylase ArnD | | Authors: | Sousa, M.C, Munoz-Escudero, D, Lee, M. | | Deposit date: | 2023-06-01 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure and Function of ArnD. A Deformylase Essential for Lipid A Modification with 4-Amino-4-deoxy-l-arabinose and Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
8T0I
 
 | |
8T0H
 
 | |
8T0G
 
 | |
8T0E
 
 | | TRPV1 in Nanodisc not bound with lysophosphatidic acid (apo) | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Cheng, Y. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8T0D
 
 | |
8T0C
 
 | |
8T0B
 
 | |
8T09
 
 | |
8T08
 
 | | Preholo-Proteasome from Pre1-1 Pre4-1 Double Mutant | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Schnell, H, Velez, B, Hanna, J. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the preholoproteasome reveals late steps in proteasome core particle biogenesis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T07
 
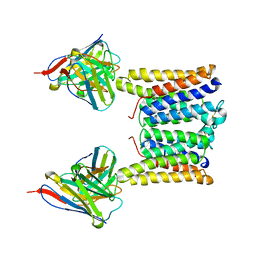 | | Structure of mouse Myomaker mutant-Y118A bound to Fab18G7 | | Descriptor: | 18G7 Fab heavy chain, 18G7 Fab light chain, Protein myomaker, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T06
 
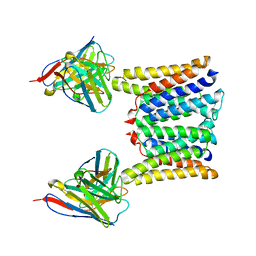 | | Structure of mouse Myomaker mutant-R107A bound to Fab18G7 | | Descriptor: | 18G7 Fab heavy chain, 18G7 Fab light chain, Protein myomaker, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T05
 
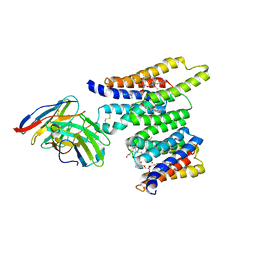 | | Structure of Ciona Myomaker bound to Fab1A1 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1A1 Fab heavy chain, 1A1 Fab light chain, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
