4OBH
 
 | |
5NN4
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-acetyl-cysteine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
4OJ4
 
 | |
6ML2
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 1) | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
5B5F
 
 | | Crystal structure of ALiS3-Streptavidin complex | | Descriptor: | N-methyl-3-(4-oxo-4,5-dihydrofuro[3,2-c]pyridin-2-yl)benzenesulfonamide, Streptavidin | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
5N1R
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4'-Pyrazol-1-ylmethyl-biphenyl-4-sulfonamide | | Descriptor: | 4-[4-(pyrazol-1-ylmethyl)phenyl]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Scozzafava, A, Carta, F. | | Deposit date: | 2017-02-06 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms
Inorg.Chim.Acta., 470, 2018
|
|
5NN5
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with 1-deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
5AR8
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biphenylsulfonamide | | Descriptor: | 2,6-bis(fluoranyl)-N-[3-[5-[2-[(3-methylsulfonylphenyl)amino]pyrimidin-4-yl]-2-morpholin-4-yl-1,3-thiazol-4-yl]phenyl]benzenesulfonamide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
4AMI
 
 | | Turkey beta1 adrenergic receptor with stabilising mutations and bound biased agonist bucindolol | | Descriptor: | 2-[(2S)-3-[[1-(1H-indol-3-yl)-2-methyl-propan-2-yl]amino]-2-oxidanyl-propoxy]benzenecarbonitrile, BETA-1 ADRENERGIC RECEPTOR, HEGA-10 | | Authors: | Warne, T, Edwards, P.C, Leslie, A.G, Tate, C.G. | | Deposit date: | 2012-03-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of a Stabilized Beta1-Adrenoceptor Bound to the Biased Agonists Bucindolol and Carvedilol
Structure, 20, 2012
|
|
4AA9
 
 | | Camel chymosin at 1.6A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHYMOSIN, GLYCEROL, ... | | Authors: | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6NO5
 
 | |
6NO2
 
 | |
3ZRJ
 
 | | Complex of ClpV N-domain with VipB peptide | | Descriptor: | 1,2-ETHANEDIOL, CLPB PROTEIN, VIPB | | Authors: | Lenherr, E.D, Kopp, J, Sinning, I. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Basis for the Unique Role of the Aaa+ Chaperone Clpv in Type Vi Protein Secretion.
J.Biol.Chem., 286, 2011
|
|
6NO0
 
 | |
5AK8
 
 | | Structure of C351A mutant of Porphyromonas gingivalis peptidylarginine deiminase | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | Authors: | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | Deposit date: | 2015-03-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
5CW6
 
 | | Structure of metal dependent enzyme DrBRCC36 | | Descriptor: | DrBRCC36, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.193 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
4A33
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 2,6-DICHLORO-4-(6-PIPERAZIN-1-YLPYRIDIN-3-YL)-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
4A30
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-BROMO-2,6-DICHLORO-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
5CLN
 
 | |
4RP7
 
 | |
5CBN
 
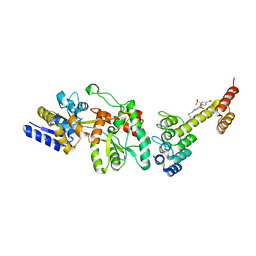 | | Fusion protein of mbp3-16 and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Maltose-binding periplasmic protein, mbp3-16,Immunoglobulin G-binding protein A | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-07-01 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
5CLO
 
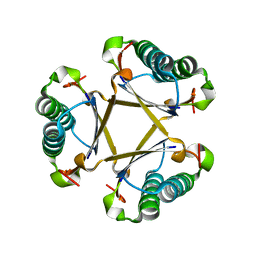 | |
4RP6
 
 | |
5CTL
 
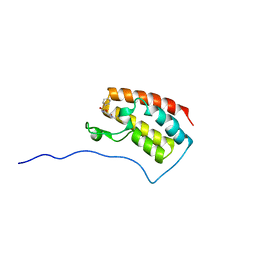 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)benzenesulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
4PDM
 
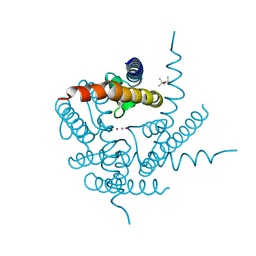 | | Crystal Structure of K+ selective NaK mutant in rubidium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, RUBIDIUM ION | | Authors: | Lam, Y. | | Deposit date: | 2014-04-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High Resolution Structural Views of Rubidium, Cesium and Barium Binding within a Potassium Selective Channel Filter
To Be Published
|
|
