4U2Q
 
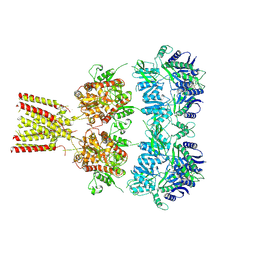 | | Full-length AMPA subtype ionotropic glutamate receptor GluA2 in complex with partial agonist kainate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5247 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U2R
 
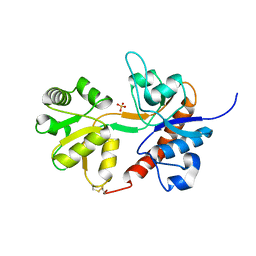 | | Crystal structure of the GLUR2 ligand binding core (S1S2J, flip variant) in the apo state | | Descriptor: | Glutamate receptor 2, SULFATE ION | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4114 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U1W
 
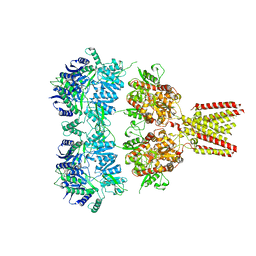 | | Full length GluA2-kainate-(R,R)-2b complex crystal form A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U1X
 
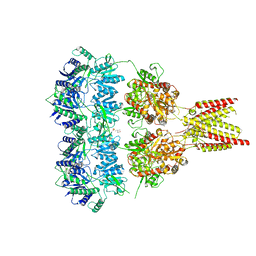 | | Full length GluA2-kainate-(R,R)-2b complex crystal form B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U1Z
 
 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form D | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9401 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U21
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form E | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3908 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U22
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form D | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4409 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U23
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form F | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6734 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U1Y
 
 | | Full length GluA2-FW-(R,R)-2b complex | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.8999 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U1O
 
 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form C | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4UQQ
 
 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQJ
 
 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQK
 
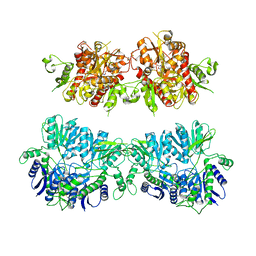 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQ6
 
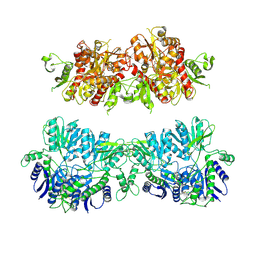 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4TLL
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 1 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
4TLM
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 2 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
4QF9
 
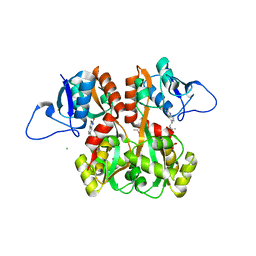 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid at 2.28 A resolution | | Descriptor: | (2S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kristensen, C.M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-05-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Binding Mode of an alpha-Amino Acid-Linked Quinoxaline-2,3-dione Analogue at Glutamate Receptor Subtype GluK1.
ACS Chem Neurosci, 6, 2015
|
|
4PE5
 
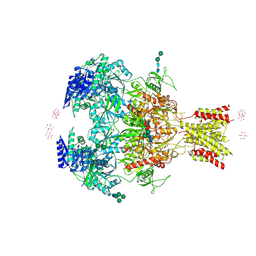 | | Crystal Structure of GluN1a/GluN2B NMDA Receptor Ion Channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol, ... | | Authors: | Karakas, E, Furukawa, H. | | Deposit date: | 2014-04-22 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Crystal structure of a heterotetrameric NMDA receptor ion channel.
Science, 344, 2014
|
|
4Q30
 
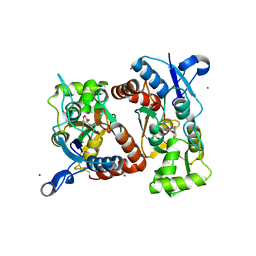 | | Nitrowillardiine bound to the ligand binding domain of GluA2 at pH 3.5 | | Descriptor: | 3-(5-nitro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-L-alanine, Glutamate receptor 2 CHIMERIC PROTEIN, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Thermodynamics and mechanism of the interaction of willardiine partial agonists with a glutamate receptor: implications for drug development.
Biochemistry, 53, 2014
|
|
4O3A
 
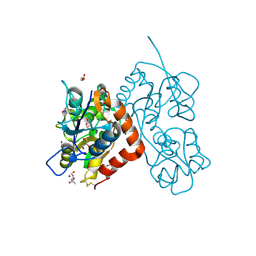 | | Crystal structure of the glua2 ligand-binding domain in complex with L-aspartate at 1.80 a resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, F, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3B
 
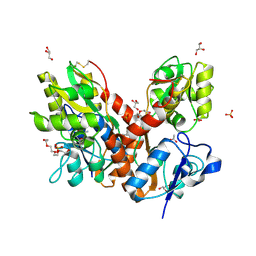 | | Crystal structure of an open/closed glua2 ligand-binding domain dimer at 1.91 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krintel, C, de Rabassa, A.C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3C
 
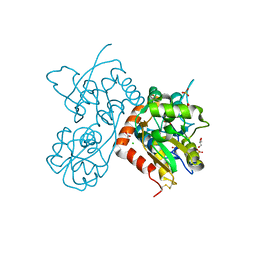 | | Crystal structure of the GLUA2 ligand-binding domain in complex with L-aspartate at 1.50 A resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, K, Kaern, A.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4NWC
 
 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | Descriptor: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4NF6
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and PPDA | | Descriptor: | (2S,3R)-1-(phenanthren-2-ylcarbonyl)piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Jespersen, A, Tajima, N, Furukawa, H. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
