6ZBI
 
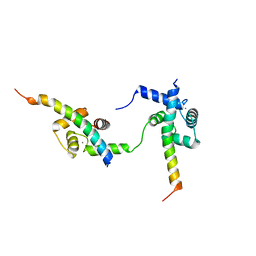 | | Ternary complex of Calmodulin bound to 2 molecules of NHE1 | | Descriptor: | CALCIUM ION, Calmodulin-1, Sodium/hydrogen exchanger 1 | | Authors: | Prestel, A, Kragelund, B.B, Pedersen, E.S, Pedersen, S.F, Sjoegaard-Frich, L.M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic Na + /H + exchanger 1 (NHE1) - calmodulin complexes of varying stoichiometry and structure regulate Ca 2+ -dependent NHE1 activation.
Elife, 10, 2021
|
|
3E89
 
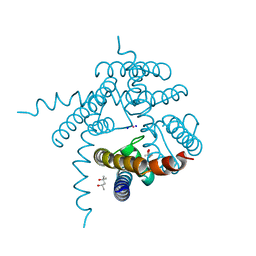 | |
3E8G
 
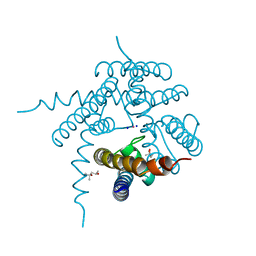 | |
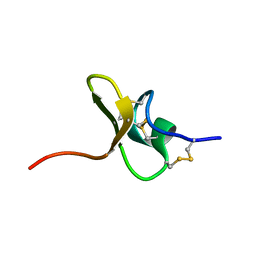
5T4R
 
 | |
7W7F
 
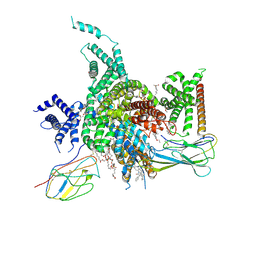 | | Cryo-EM structure of human NaV1.3/beta1/beta2-ICA121431 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-04 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
7W77
 
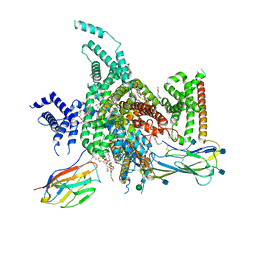 | | cryo-EM structure of human NaV1.3/beta1/beta2-bulleyaconitineA | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
2LT9
 
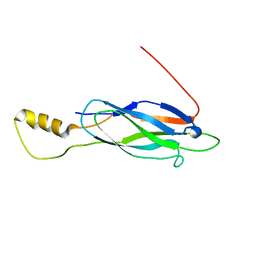 | |
9BWT
 
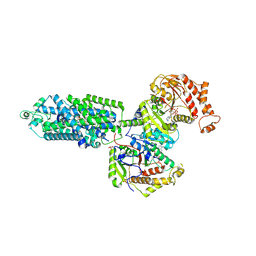 | | human sodium chloride cotransporter NCC S344E in the phosphorylation state and in complex with hydrochlorothiazide | | Descriptor: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, ADENOSINE-5'-TRIPHOSPHATE, Solute carrier family 12 member 3 | | Authors: | Cao, E.H, Zhao, Y.X. | | Deposit date: | 2024-05-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural bases for Na + -Cl - cotransporter inhibition by thiazide diuretic drugs and activation by kinases.
Nat Commun, 15, 2024
|
|
9DBM
 
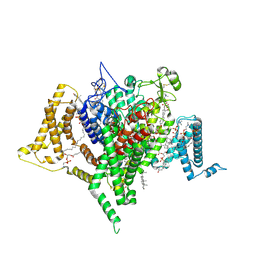 | | Full-length apo human voltage-gated sodium channel 1.8 (NaV1.8), class II | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Neumann, B, McCarthy, S, Gonen, S. | | Deposit date: | 2024-08-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of inhibition of human Na V 1.8 by the tarantula venom peptide Protoxin-I.
Nat Commun, 16, 2025
|
|
9DBN
 
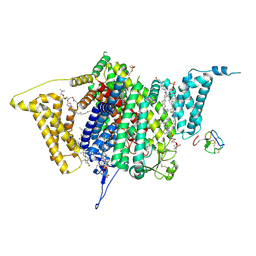 | | Tarantula venom peptide Protoxin-I bound to full-length human voltage-gated sodium channel 1.8 (NaV1.8) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Beta/omega-theraphotoxin-Tp1a, ... | | Authors: | Neumann, B, McCarthy, S, Gonen, S. | | Deposit date: | 2024-08-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of inhibition of human Na V 1.8 by the tarantula venom peptide Protoxin-I.
Nat Commun, 16, 2025
|
|
9DBL
 
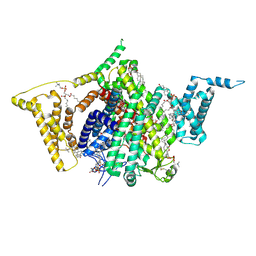 | | Full-length apo human voltage-gated sodium channel 1.8 (NaV1.8), class I | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Neumann, B, McCarthy, S, Gonen, S. | | Deposit date: | 2024-08-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural basis of inhibition of human Na V 1.8 by the tarantula venom peptide Protoxin-I.
Nat Commun, 16, 2025
|
|
9DBK
 
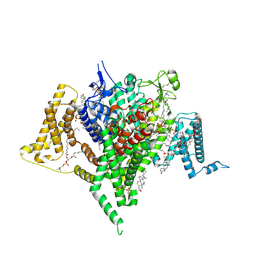 | | Full-length apo human voltage-gated sodium channel 1.8 (NaV1.8) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Neumann, B, McCarthy, S, Gonen, S. | | Deposit date: | 2024-08-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural basis of inhibition of human Na V 1.8 by the tarantula venom peptide Protoxin-I.
Nat Commun, 16, 2025
|
|
7NGH
 
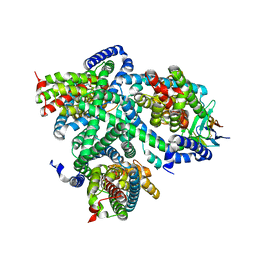 | | Structure of glutamate transporter homologue in complex with Sybody | | Descriptor: | ASPARTIC ACID, Proton/glutamate symporter, SDF family, ... | | Authors: | Arkhipova, V, Slotboom, D.J, Guskov, A. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Kinetic mechanism of Na + -coupled aspartate transport catalyzed by Glt Tk .
Commun Biol, 4, 2021
|
|
7YS5
 
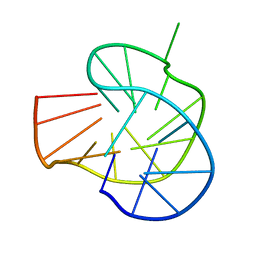 | | RET G-quadruplex in 10mM Na+ | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Yin, S, Cao, C. | | Deposit date: | 2022-08-11 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RET G-quadruplex in 10mM Na+
To Be Published
|
|
4RKO
 
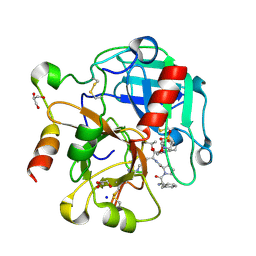 | | Crystal structure of thrombin mutant S195T bound with PPACK | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | Deposit date: | 2014-10-13 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
1A5U
 
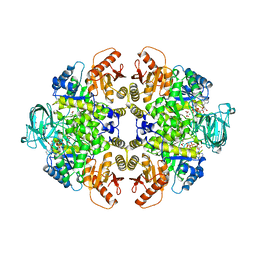 | | PYRUVATE KINASE COMPLEX WITH BIS MG-ATP-NA-OXALATE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Larsen, T.M, Benning, M.M, Rayment, I, Reed, G.H. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the bis(Mg2+)-ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 A resolution: ATP binding over a barrel.
Biochemistry, 37, 1998
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
5A1S
 
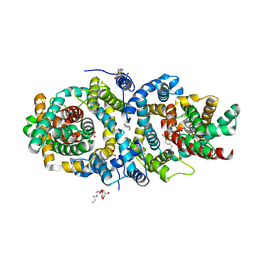 | | Crystal structure of the sodium-dependent citrate symporter SeCitS form Salmonella enterica. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Woehlert, D, Groetzinger, M.J, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2015-05-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Na(+)-dependent citrate transport from the structure of an asymmetrical CitS dimer.
Elife, 4, 2015
|
|
5F1F
 
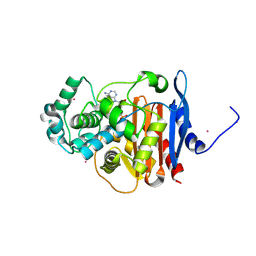 | | Crystal structure of CMY-10 adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, CADMIUM ION | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
1HXE
 
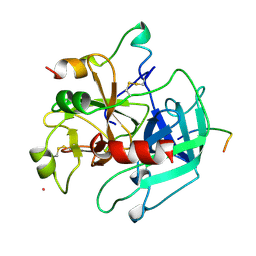 | | SERINE PROTEASE | | Descriptor: | HIRUDIN VARIANT-1, RUBIDIUM ION, THROMBIN | | Authors: | Tulinsky, A, Zhang, E. | | Deposit date: | 1995-12-07 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular environment of the Na+ binding site of thrombin.
Biophys.Chem., 63, 1997
|
|
7WFW
 
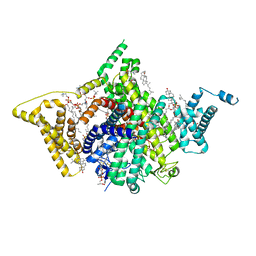 | | Apo human Nav1.8 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2FMJ
 
 | |
7WFR
 
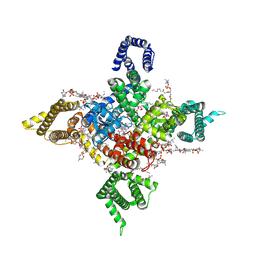 | | Human Nav1.8 with A-803467, class III | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WEL
 
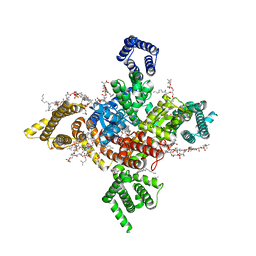 | | Human Nav1.8 with A-803467, class II | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WE4
 
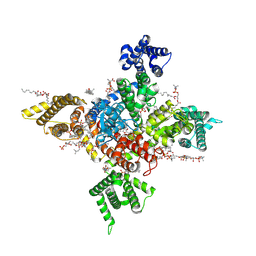 | | Human Nav1.8 with A-803467, class I | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
