8VAT
 
 | |
8VAN
 
 | | Structure of the E. coli clamp loader bound to the beta clamp in an Initial-Binding conformation | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Landeck, J.T, Pajak, J, Kelch, B.A. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Differences between bacteria and eukaryotes in clamp loader mechanism, a conserved process underlying DNA replication.
J.Biol.Chem., 300, 2024
|
|
8VAR
 
 | |
8VAL
 
 | |
8VAQ
 
 | |
8VAP
 
 | |
8VAM
 
 | |
8VAS
 
 | |
7LDD
 
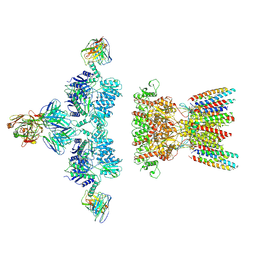 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LDE
 
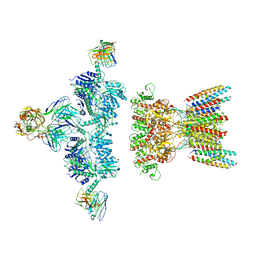 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
2JOS
 
 | |
9C3F
 
 | | Cryo-EM structure of E. coli AmpG | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Anhydromuropeptide permease,Soluble cytochrome b562, DODECYL-BETA-D-MALTOSIDE | | Authors: | Sverak, H, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2024-05-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM characterization of the anydromuropeptide permease AmpG central to bacterial fitness and beta-lactam antibiotic resistance.
Nat Commun, 15, 2024
|
|
8Y55
 
 | | Crystal Structure of NAMPT-PF403 | | Descriptor: | (13~{a}~{S})-6,7-dimethoxy-9,11,12,13,13~{a},14-hexahydrophenanthro[9,10-f]indolizin-3-ol, DI(HYDROXYETHYL)ETHER, Nicotinamide phosphoribosyltransferase | | Authors: | Yu, S.S, Liu, Y.B, Li, Y. | | Deposit date: | 2024-01-31 | | Release date: | 2025-02-19 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Thermal proteome profiling (TPP) reveals NAMPT as the anti-glioma target of phenanthroindolizidine alkaloid PF403.
Acta Pharm Sin B, 15, 2025
|
|
8ATH
 
 | | CRYSTAL STRUCTURE OF LAMP1 IN COMPLEX WITH FAB-B. | | Descriptor: | Fab B Heavy Chain, Fab B Light Chain, Lysosome-associated membrane glycoprotein 1 | | Authors: | Mathieu, M, Dupuy, A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.366 Å) | | Cite: | Deciphering cross-species reactivity of LAMP-1 antibodies using deep mutational epitope mapping and AlphaFold.
Mabs, 15, 2023
|
|
8FYF
 
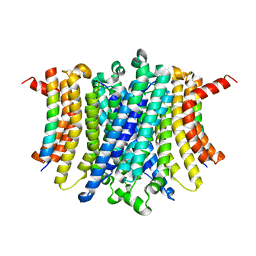 | | Human TMEM175-LAMP1 transmembrane domain only complex | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, Lysosome-associated membrane glycoprotein 1 | | Authors: | Zhang, J.Y, Zeng, W.Z, Han, Y, Jiang, Y.X. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel.
Mol.Cell, 83, 2023
|
|
8FY5
 
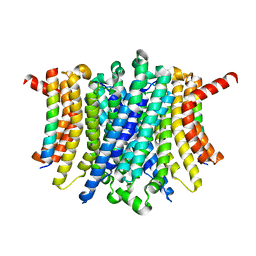 | | Human TMEM175-LAMP1 full-length complex | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, Lysosome-associated membrane glycoprotein 1 | | Authors: | Zhang, J.Y, Zeng, W.Z, Han, Y, Jiang, Y.X. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel.
Mol.Cell, 83, 2023
|
|
7Q8T
 
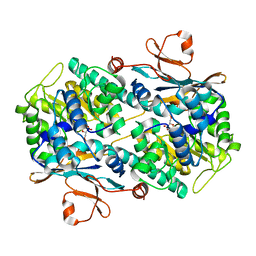 | | Crystal structure of NAMPT bound to ligand TSY535(compound 9a) | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION, [(2~{R},3~{S},4~{R},5~{S})-3,4-bis(oxidanyl)-5-[4-[[[4-(phenylsulfonyl)phenyl]carbamoylamino]methyl]phenyl]oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Kraemer, A, Tang, S, Butterworth, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Chemistry-led investigations into the mode of action of NAMPT activators, resulting in the discovery of non-pyridyl class NAMPT activators.
Acta Pharm Sin B, 13, 2023
|
|
8YJX
 
 | |
7PPH
 
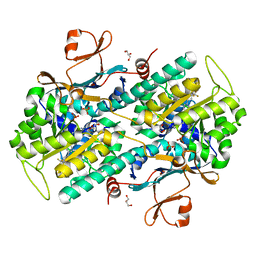 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH Compound 10 | | Descriptor: | GLYCEROL, N-[4-[(5R)-6-oxidanylidene-5-quinolin-5-yl-5-(trifluoromethyl)-1,4-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
7PPI
 
 | | Crystal STRUCTURE OF NAMPT IN COMPLEX WITH Compound 11 | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-[(5R)-1-(4-azanylbutyl)-6-oxidanylidene-5-quinolin-5-yl-4,5-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
7PPE
 
 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH COMPOUND 1 | | Descriptor: | GLYCEROL, N-[4-[(4R)-4-methyl-1-(oxan-4-yl)-6-oxidanylidene-4,5-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
7PPF
 
 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH COMPOUND 8 | | Descriptor: | 1,2-ETHANEDIOL, N-[4-[(4R)-1,4-dimethyl-6-oxidanylidene-4,5-dihydropyridazin-3-yl]phenyl]-5,7-dihydropyrrolo[3,4-b]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
7PPG
 
 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 1,2-ETHANEDIOL, N-[4-[(4R)-1-cyclopentyl-4-methyl-6-oxidanylidene-4,5-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
5FXT
 
 | |
6YEO
 
 | | Crystal structure of AmpC from E. coli with cyclic boronate 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Lang, P.A, Schofield, C.J, Brem, J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
