6TRO
 
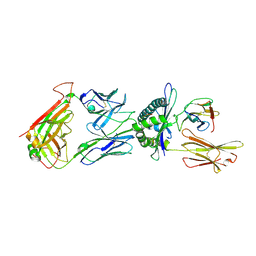 | | Crystal structure of the T-cell receptor GVY01 bound to HLA A2*01-GVYDGREHTV | | Descriptor: | Beta-2-microglobulin, MAGE-A4 peptide (amino acids 230-239), MHC class I antigen, ... | | Authors: | Coles, C.H, McMurran, C, Lloyd, A, Hibbert, L, Lupardus, P.J, Cole, D.K, Harper, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
J.Biol.Chem., 295, 2020
|
|
6IYC
 
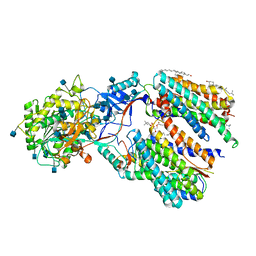 | | Recognition of the Amyloid Precursor Protein by Human gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, R, Yang, G, Guo, X, Zhou, Q, Lei, J, Shi, Y. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of the amyloid precursor protein by human gamma-secretase.
Science, 363, 2019
|
|
5LJ3
 
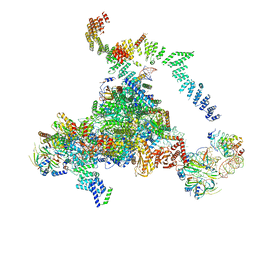 | | Structure of the core of the yeast spliceosome immediately after branching | | Descriptor: | CEF1, CLF1, CWC15, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-03 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5LQW
 
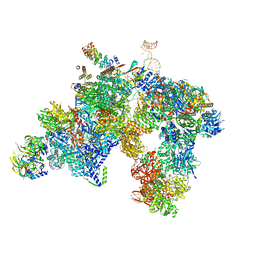 | | yeast activated spliceosome | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-processing protein 45, Pre-mRNA-splicing factor 8, ... | | Authors: | Rauhut, R, Luehrmann, R. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Molecular architecture of the Saccharomyces cerevisiae activated spliceosome
Science, 6306, 2016
|
|
4UIS
 
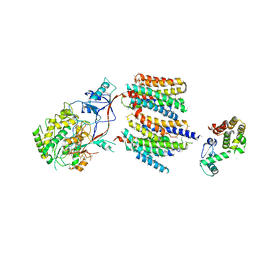 | | The cryoEM structure of human gamma-Secretase complex | | Descriptor: | GAMMA-SECRETASE, LYSOZYME | | Authors: | Sun, L, Zhao, L, Yang, G, Yan, C, Zhou, R, Zhou, X, Xie, T, Zhao, Y, Wu, S, Li, X, Shi, Y. | | Deposit date: | 2015-04-03 | | Release date: | 2015-06-10 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis of Human Gamma-Secretase Assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6TRN
 
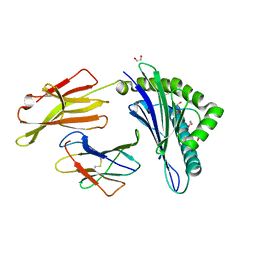 | | Crystal structure of HLA A2*01-AVYDGREHTV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MAGE-A4 peptide (amino acids 230-239) variant, ... | | Authors: | Coles, C.H, McMurran, C, Lloyd, A, Hibbert, L, Lupardus, P.J, Cole, D.K, Harper, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
J.Biol.Chem., 295, 2020
|
|
3VS8
 
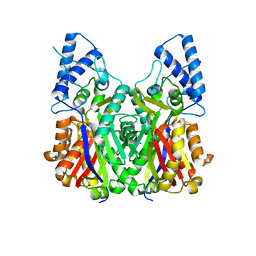 | | Crystal structure of type III PKS ArsC | | Descriptor: | SODIUM ION, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
5MPS
 
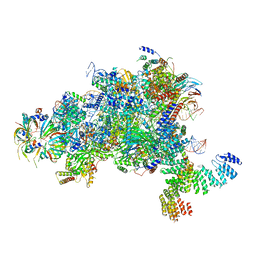 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5N4B
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 25mer macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
3VS9
 
 | | Crystal structure of type III PKS ArsC mutant | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
3WKH
 
 | | Crystal structure of cellobiose 2-epimerase in complex with epilactose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKG
 
 | | Crystal structure of cellobiose 2-epimerase in complex with glucosylmannose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKI
 
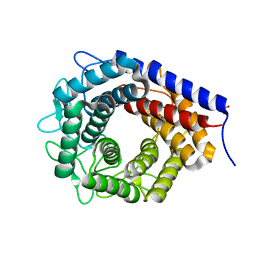 | | Crystal structure of cellobiose 2-epimerase in complex with cellobiitol | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKF
 
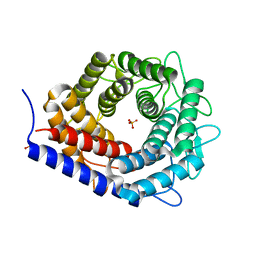 | | Crystal structure of cellobiose 2-epimerase | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
7FIM
 
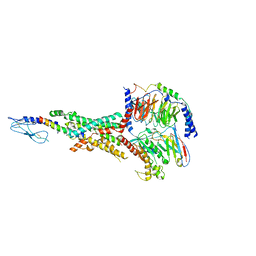 | | Cryo-EM structure of the tirzepatide (LY3298176)-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,human glucagon like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-07-31 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
4V88
 
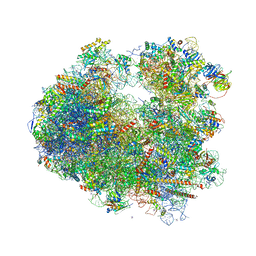 | | The structure of the eukaryotic ribosome at 3.0 A resolution. | | Descriptor: | 18S RIBOSOMAL RNA, 18S rRNA, 25S rRNA, ... | | Authors: | Ben-Shem, A, Garreau de Loubresse, N, Melnikov, S, Jenner, L, Yusupova, G, Yusupov, M. | | Deposit date: | 2011-10-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the eukaryotic ribosome at 3.0 angstrom resolution.
Science, 334, 2011
|
|
4V92
 
 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 18S RRNA, ES1, ES10, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
8IM5
 
 | |
4W97
 
 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4WCJ
 
 | | Structure of IcaB from Ammonifex degensii | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
7T8F
 
 | |
7T8E
 
 | | G93A mutant of human SOD1 in P21 space group | | Descriptor: | ACETATE ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Amporndanai, K, Hasnain, S.S. | | Deposit date: | 2021-12-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ebselen analogues delay disease onset and its course in fALS by on-target SOD-1 engagement.
Sci Rep, 14, 2024
|
|
7F24
 
 | |
