6NAK
 
 | | BACTERIAL PROTEIN COMPLEX TM BDE complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, TsaE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-05 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
6MNT
 
 | |
6MK7
 
 | | Solution structure of the large extracellular loop of FtsX in Streptococcus pneumoniae | | Descriptor: | Cell division protein FtsX | | Authors: | Edmonds, K.A, Fu, Y, Wu, H, Rued, B.E, Bruce, K.E, Winkler, M.E, Giedroc, D.P. | | Deposit date: | 2018-09-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
MBio, 10, 2019
|
|
6STX
 
 | | Copper oxidase from Colletotrichum graminicola | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Kelch domain-containing protein, ... | | Authors: | Offen, W.A, Henrissat, B, Davies, G.J. | | Deposit date: | 2019-09-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Fungal Copper Radical Oxidase with High Catalytic Efficiency toward 5-Hydroxymethylfurfural and Benzyl Alcohols for Bioprocessing
Acs Catalysis, 2020
|
|
6MV3
 
 | | NMR structure of the cNTnC-cTnI chimera bound to calcium desensitizer W7 | | Descriptor: | CALCIUM ION, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE, Troponin C, ... | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2018-10-24 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Changes Induced by the Binding of the Calcium Desensitizer W7 to Cardiac Troponin.
Biochemistry, 57, 2018
|
|
6MIF
 
 | | Lim5 domain of PINCH1 protein | | Descriptor: | LIM and senescent cell antigen-like-containing domain protein 1, ZINC ION | | Authors: | Qin, J, Vaynberg, J. | | Deposit date: | 2018-09-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Non-catalytic signaling by pseudokinase ILK for regulating cell adhesion.
Nat Commun, 9, 2018
|
|
4W6Q
 
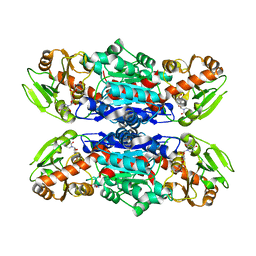 | | Glycosyltransferase C from Streptococcus agalactiae | | Descriptor: | SERINE, URIDINE-5'-DIPHOSPHATE, glucosyltransferase | | Authors: | Zhu, F, Zhang, H, Wu, H. | | Deposit date: | 2014-08-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved domain is crucial for acceptor substrate binding in a family of glucosyltransferases.
J.Bacteriol., 197, 2015
|
|
4W74
 
 | |
6MY2
 
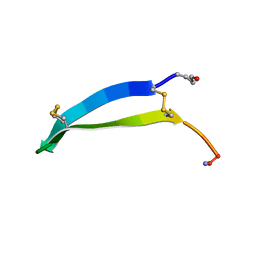 | | Solution structure of gomesin at 298 K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
6MJV
 
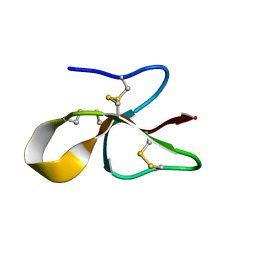 | | A consensus human beta defensin | | Descriptor: | Human beta-defensin | | Authors: | Amero, C, Villegas-Moreno, J, Rodriguez, A, Norton, R.S, Corzo, G. | | Deposit date: | 2018-09-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial activity and structure of a consensus human beta-defensin and its comparison to a novel putative hBD10.
Proteins, 88, 2020
|
|
6MY1
 
 | | Solution structure of gomesin at 278 K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
4W7Y
 
 | |
6MWM
 
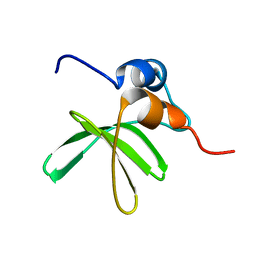 | | Bat coronavirus HKU4 SUD-C | | Descriptor: | Non-structural protein 3 | | Authors: | Staup, A.J, De Silva, I.U, Catt, J.T, Tan, X, Hammond, R.G, Johnson, M.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the SARS-Unique Domain C From the Bat Coronavirus HKU4.
Nat Prod Commun, 14, 2019
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MK5
 
 | |
6MM4
 
 | |
4W8T
 
 | |
4W8Z
 
 | |
6MNL
 
 | |
5LGK
 
 | | Crystal structure of the human IgE-Fc bound to its B cell receptor derCD23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig epsilon chain C region, Low affinity immunoglobulin epsilon Fc receptor, ... | | Authors: | Dhaliwal, B, Pang, M.O.Y, Sutton, B.J. | | Deposit date: | 2016-07-07 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | IgE binds asymmetrically to its B cell receptor CD23.
Sci Rep, 7, 2017
|
|
6NAN
 
 | | NMR structure determination of Ixolaris and Factor X interaction reveals a noncanonical mechanism of Kunitz inhibition | | Descriptor: | Ixolaris | | Authors: | De Paula, V.S, Sgourakis, N.G, Francischetti, I.M.B, Almeida, F.C.L, Monteiro, R.Q, Valente, A.P. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of Ixolaris and factor X(a) interaction reveals a noncanonical mechanism of Kunitz inhibition.
Blood, 134, 2019
|
|
6MY3
 
 | | Solution structure of gomesin at 310K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
6MZT
 
 | | Solution structure of alpha-KTx-6.21 (UroTx) from Urodacus yaschenkoi | | Descriptor: | Potassium channel toxin alpha-KTx 6.21 | | Authors: | Chin, Y.K.-Y, Luna-Ramirez, K, Anangi, R, King, G.F. | | Deposit date: | 2018-11-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the potency and selectivity of Urotoxin, a potent Kv1 blocker from scorpion venom.
Biochem. Pharmacol., 174, 2020
|
|
6NW8
 
 | | SOLUTION STRUCTURE OF CN29, A TOXIN FROM CENTRUROIDES NOXIUS SCORPION VENOM | | Descriptor: | Cn29 | | Authors: | Delepierre, M, Gurrola, G.B, Possani, L.D, Guijarro, J.I. | | Deposit date: | 2019-02-06 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Cn29, a novel orphan peptide found in the venom of the scorpion Centruroides noxius: Structure and function.
Toxicon, 167, 2019
|
|
4WA8
 
 | |
