8HIO
 
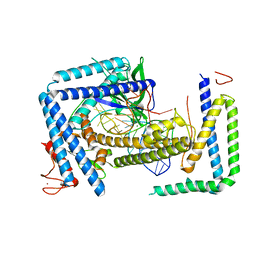 | | Cryo-EM structure of the Cas12m2-crRNA binary complex | | Descriptor: | Cas12m2, MAGNESIUM ION, RNA (56-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HHM
 
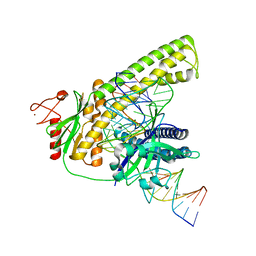 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA ternary complex intermediate state | | Descriptor: | Cas12m2, DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5F9H
 
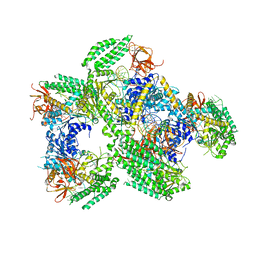 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer 5' triphosphate hairpin RNA | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8I43
 
 | |
8I44
 
 | |
8I45
 
 | |
8I46
 
 | |
8TNS
 
 | |
7TQV
 
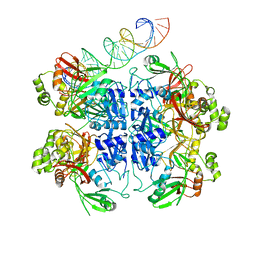 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (33-MER), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
7TJ2
 
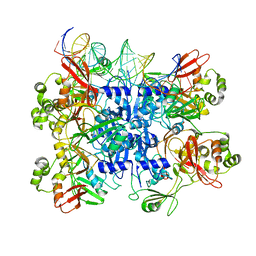 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (31-MER), Uridylate-specific endoribonuclease nsp15 | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
6I1W
 
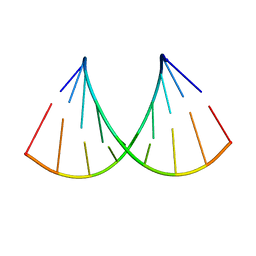 | | Structure of the RNA duplex containing pseudouridine residue (5'-Gp(PSU)pC-3' sequence context) | | Descriptor: | RNA (5'-R(*AP*CP*UP*GP*AP*CP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*GP*(PSU)P*CP*AP*GP*U)-3') | | Authors: | Deb, I, Popenda, L, Sarzynska, J, Gdaniec, Z. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair.
Sci Rep, 9, 2019
|
|
6I1V
 
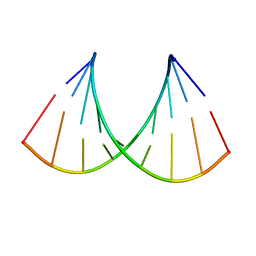 | | Structure of the RNA duplex containing pseudouridine residue (5'-Cp(PSU)pG-3' sequence context) | | Descriptor: | RNA (5'-R(*AP*CP*UP*CP*AP*GP*UP*GP*A)-3'), RNA (5'-R(*UP*CP*AP*CP*(PSU)P*GP*AP*GP*U)-3') | | Authors: | Deb, I, Popenda, L, Sarzynska, J, Gdaniec, Z. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational and NMR studies of RNA duplexes with an internal pseudouridine-adenosine base pair.
Sci Rep, 9, 2019
|
|
1PUD
 
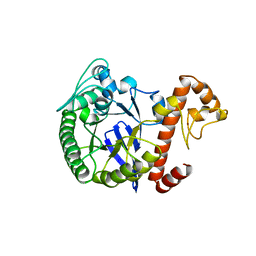 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-06-28 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange.
EMBO J., 15, 1996
|
|
6PIG
 
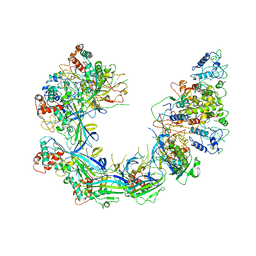 | | V. cholerae TniQ-Cascade complex, closed conformation | | Descriptor: | RNA (60-MER), TniQ monomer 1, TniQ monomer 2, ... | | Authors: | Halpin-Healy, T, Klompe, S, Sternberg, S.H. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system.
Nature, 577, 2020
|
|
8DMB
 
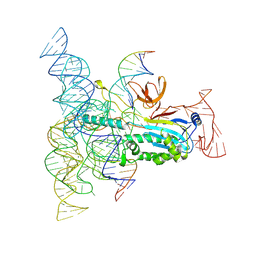 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | Descriptor: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | Authors: | Seiichi, H, Kappel, K, Zhang, F. | | Deposit date: | 2022-07-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
1PXG
 
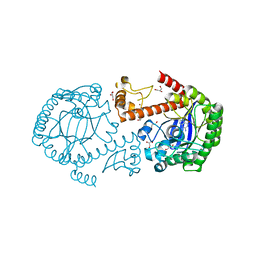 | | Crystal structure of the mutated tRNA-guanine transglycosylase (TGT) D280E complexed with preQ1 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Kittendorf, J.D, Sgraja, T, Reuter, K, Klebe, G, Garcia, G.A. | | Deposit date: | 2003-07-04 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An essential role for aspartate 264 in catalysis by tRNA-guanine transglycosylase from Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
3TP2
 
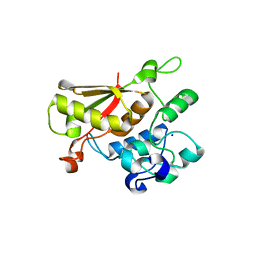 | | Crystal Structure of the Splicing Factor Cwc2 from yeast | | Descriptor: | Pre-mRNA-splicing factor CWC2, SODIUM ION, ZINC ION | | Authors: | Schmitzova, J. | | Deposit date: | 2011-09-07 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein.
Embo J., 31, 2012
|
|
1OXJ
 
 | |
8HHL
 
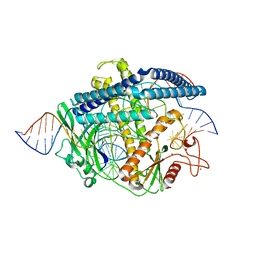 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA full R-loop complex | | Descriptor: | Cas12m2, MAGNESIUM ION, NTS (36-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6OV0
 
 | |
7TDB
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine bisphosphonate, manganese ions | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, [2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethoxy-oxidanyl-phosphoryl]methylphosphonic acid, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery.
Acs Chem.Biol., 17, 2022
|
|
7TD7
 
 | |
7TDC
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine bisphosphonate, calcium ions | | Descriptor: | CALCIUM ION, MAGNESIUM ION, [2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethoxy-oxidanyl-phosphoryl]methylphosphonic acid, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Subsite Ligand Recognition and Cooperativity in the TPP Riboswitch: Implications for Fragment-Linking in RNA Ligand Discovery.
Acs Chem.Biol., 17, 2022
|
|
7TDA
 
 | |
7V94
 
 | | Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex | | Descriptor: | Cas12c2, sgRNA, target DNA (non target strand), ... | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
