7WAY
 
 | |
7WB1
 
 | |
3KMN
 
 | | Crystal Structure of the Human Apo GST Pi C47S/Y108V Double Mutant | | Descriptor: | CALCIUM ION, CARBONATE ION, Glutathione S-transferase P, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diuretic drug binding to human glutathione transferase P1-1: potential role of CYS101 revealed in the double mutant C47S/Y108V
to be published
|
|
1MHD
 
 | | CRYSTAL STRUCTURE OF A SMAD MH1 DOMAIN BOUND TO DNA | | Descriptor: | DNA, SMAD3 | | Authors: | Shi, Y. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling.
Cell(Cambridge,Mass.), 94, 1998
|
|
6QG4
 
 | |
6QFT
 
 | | Structure of the mitogen activated kinase kinase 7 in complex with pyrazolopyrimidin 1b | | Descriptor: | 1-[(3~{R})-3-(4-azanyl-3-iodanyl-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Wolle, P, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Covalent Pyrazolopyrimidine-MKK7 Complexes and a Report on a Unique DFG-in/Leu-in Conformation of Mitogen-Activated Protein Kinase Kinase 7 (MKK7).
J.Med.Chem., 62, 2019
|
|
6QFL
 
 | |
6QG7
 
 | | Structure of the mitogen activated kinase kinase 7 in complex with pyrazolopyrimidine 1k | | Descriptor: | (R)-1-(3-(4-amino-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Wolle, P, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of Covalent Pyrazolopyrimidine-MKK7 Complexes and a Report on a Unique DFG-in/Leu-in Conformation of Mitogen-Activated Protein Kinase Kinase 7 (MKK7).
J.Med.Chem., 62, 2019
|
|
6QHR
 
 | | Structure of the mitogen activated kinase kinase 7 in complex with pyrazolopyrimidine 1m | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Wolle, P, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Characterization of Covalent Pyrazolopyrimidine-MKK7 Complexes and a Report on a Unique DFG-in/Leu-in Conformation of Mitogen-Activated Protein Kinase Kinase 7 (MKK7).
J.Med.Chem., 62, 2019
|
|
6QHO
 
 | |
6QFR
 
 | |
6R7I
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R6H
 
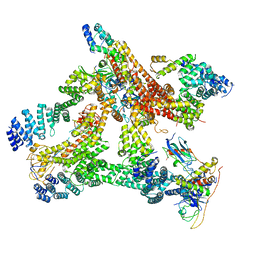 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Morris, E.P, Faull, S.V, Lau, A.M.C, Politis, A, Beuron, F, Cronin, N. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7H
 
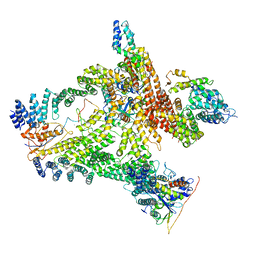 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
1MD4
 
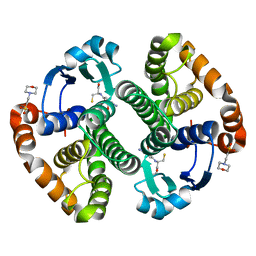 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to valine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
6R7F
 
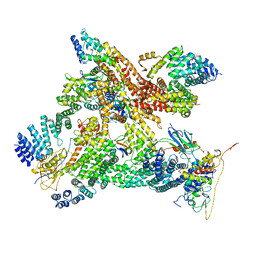 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
1MD3
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to alanine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
3KM6
 
 | |
1LBK
 
 | | Crystal structure of a recombinant glutathione transferase, created by replacing the last seven residues of each subunit of the human class pi isoenzyme with the additional C-terminal helix of human class alpha isoenzyme | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase class pi chimaera (CODA), ... | | Authors: | Kong, G.K.W, Micaloni, C, Mazzetti, A.P, Nuccetelli, M, Antonini, G, Stella, L, McKinstry, W.J, Polekhina, G, Rossjohn, J, Federici, G, Ricci, G, Parker, M.W, Lo Bello, M. | | Deposit date: | 2002-04-04 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering a new C-terminal tail in the H-site of human glutathione transferase P1-1: structural and functional consequences.
J.Mol.Biol., 325, 2003
|
|
3KMO
 
 | |
8AFK
 
 | | Structure of iRFP variant C15S/N136R/V256C in complex with phycocyanobilin | | Descriptor: | Near-infrared fluorescent protein, PHYCOCYANOBILIN | | Authors: | Remeeva, A, Kovalev, K, Gushchin, I, Fonin, A, Turoverov, K, Stepanenko, O. | | Deposit date: | 2022-07-18 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structure of iRFP variant C15S/N136R/V256C in complex with phycocyanobilin
To Be Published
|
|
8AM0
 
 | | Crystal structure of human T1061E PI3Kalpha in complex with its regulatory subunit and the inhibitor GDC-0077 (Inavolisib) | | Descriptor: | (2R)-2-[[2-[(4S)-4-[bis(fluoranyl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl]amino]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Goncalves, M, Johnson, J.L, Roewer, K.M. | | Deposit date: | 2022-08-02 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Epinephrine inhibits PI3K alpha via the Hippo kinases.
Cell Rep, 42, 2023
|
|
8C17
 
 | | Crystal structure of TEAD4 in complex with peptide 1 | | Descriptor: | GLYCEROL, MYRISTIC ACID, Stapled peptide, ... | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical and Structural Characterization of a Peptidic Inhibitor of the YAP:TEAD Interaction That Binds to the alpha-Helix Pocket on TEAD.
Acs Chem.Biol., 18, 2023
|
|
8CAA
 
 | | Crystal structure of TEAD4 in complex with YTP-13 | | Descriptor: | (2~{R})-2-[2-chloranyl-5-[2-chloranyl-4-(trifluoromethyl)phenoxy]phenyl]sulfanylpropanoic acid, 4-[bis(fluoranyl)methoxy]-2-[(2~{S})-5-chloranyl-6-fluoranyl-2-[[(4-oxidanylcyclohexyl)amino]methyl]-2-phenyl-3~{H}-1-benzofuran-4-yl]-3-fluoranyl-benzamide, PHOSPHATE ION, ... | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Optimization of a Class of Dihydrobenzofurane Analogs toward Orally Efficacious YAP-TEAD Protein-Protein Interaction Inhibitors.
Chemmedchem, 18, 2023
|
|
8CK3
 
 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (S)-1-(3,5-Difluoro-phenyl)-5,5-difluoro-3-methanesulfonyl-5,6-dihydro-4H-cyclopenta[c]thiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-4,6-dihydrocyclopenta[c]thiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Lehmannn, M, Diehl, L. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
